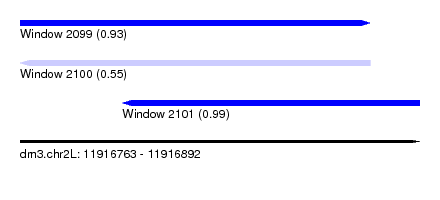
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,916,763 – 11,916,892 |
| Length | 129 |
| Max. P | 0.989146 |

| Location | 11,916,763 – 11,916,876 |
|---|---|
| Length | 113 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.32 |
| Shannon entropy | 0.34787 |
| G+C content | 0.42269 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -22.09 |
| Energy contribution | -22.21 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11916763 113 + 23011544 ------UAAACGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACAGGGCGGACAUCGGGUGGAUAUAUCUACAAUACUG ------....((((....(((((((((((.(((........)))))).))))))))(..((((((((.....))))))))..)...))))........(((((....)))))....... ( -28.30, z-score = -2.59, R) >droSim1.chr2L 11723961 113 + 22036055 ------UAAACGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACAGGUCGGACAUCGGGUGGAUAUAUCCACUAUAUUG ------....((((....(((((((((((.(((........)))))).))))))))(..((((((((.....))))))))..)..)).)).......((((((....))))))...... ( -28.00, z-score = -2.59, R) >droSec1.super_16 116095 113 + 1878335 ------UAAACGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCAACAGGGCGGACAUCGGGUGGAUAUAUCCACUAUAUUG ------....((((....(((((((((((.(((........)))))).))))))))...((((((((.....))))))))......)))).......((((((....))))))...... ( -31.30, z-score = -3.81, R) >droYak2.chr2L 8336663 112 + 22324452 ------UAAACGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACAGGGCGGACAUCGGCUGGAUCUA-CUACUAUACUU ------.....(((((.....)).......(((.((((((..((.......))......((((((((.....)))))))).))))))...)))...)))........-........... ( -24.30, z-score = -1.25, R) >droEre2.scaffold_4929 13144297 107 - 26641161 ------UAAACGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACAGGGCGGACAUCGGCUGGAUAUA-CUACUG----- ------....((((....(((((((((((.(((........)))))).))))))))(..((((((((.....))))))))..)...)))).....(((.(((.....-))))))----- ( -24.80, z-score = -1.48, R) >droAna3.scaffold_12943 2689681 97 - 5039921 ------UAAACGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACAGGGCGGACAUCACAGCGA---------------- ------....((((....(((((((((((.(((........)))))).))))))))(..((((((((.....))))))))..)...)))).............---------------- ( -24.20, z-score = -2.30, R) >dp4.chr4_group3 9236871 109 + 11692001 ------UGAGCGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACGGGGCGAGC-CGGGCUGCAGGA---UGCUGGACAG ------.....((((((((((((....((((((.((.(((..((.......))......((((((((.....)))))))).))))))))))).-......))))))---)).))).).. ( -31.10, z-score = -0.37, R) >droPer1.super_8 428115 109 + 3966273 ------UGAGCGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACGGGGCGGGC-CCGGCUGCAGGA---UGCUGGACAG ------.....((((((((((((((((((.(((........)))))))))(((...(..((((((((.....))))))))..)((((.....)-))))))))))))---)).))).).. ( -33.70, z-score = -0.96, R) >droWil1.scaffold_180708 2601910 119 + 12563649 GUAGAAAAAAUGCGGCAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGAUGGGGCGGAGGCUAGUGGUUGAUAGCCUGAACAACG ((((((((((((...)))))).)))))).......(((((((((.((....)).(((((((......(((((((.....)))))))(((....))))))))))....))))).)))).. ( -29.50, z-score = -0.42, R) >consensus ______UAAACGCCACAUUUUGUUUUACUCGUCACUUGUCAGGCAGUAAAAGCAAAUUACUUAAAGUUCAUUGCUUUAAGCGACAGGGCGGACAUCGGCUGGAUAUA_CUACUAUACUG ..........((((....(((((((((((.(((........)))))).))))))))...((((((((.....))))))))......))))............................. (-22.09 = -22.21 + 0.12)

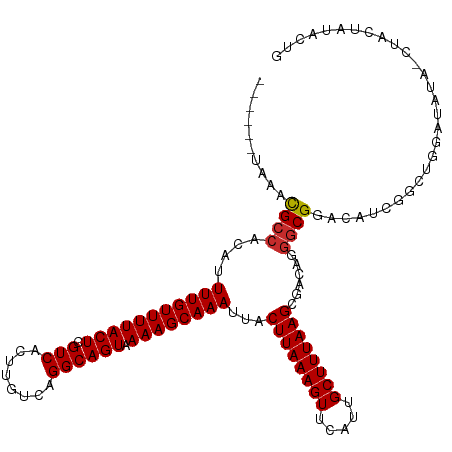
| Location | 11,916,763 – 11,916,876 |
|---|---|
| Length | 113 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.32 |
| Shannon entropy | 0.34787 |
| G+C content | 0.42269 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.33 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11916763 113 - 23011544 CAGUAUUGUAGAUAUAUCCACCCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGUUUA------ ..........(((((.........)))))((((.....((((((((.......))))))))..((((((((((((........))))).))))))).........))))....------ ( -22.20, z-score = -0.70, R) >droSim1.chr2L 11723961 113 - 22036055 CAAUAUAGUGGAUAUAUCCACCCGAUGUCCGACCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGUUUA------ .......(((((....)))))..((((((((...(((.((((((((.......))))))))..((((((((((((........))))).))))))))))...)).))))))..------ ( -25.70, z-score = -1.82, R) >droSec1.super_16 116095 113 - 1878335 CAAUAUAGUGGAUAUAUCCACCCGAUGUCCGCCCUGUUGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGUUUA------ .......(((((....)))))..((((.((((....((((((((((.......))))))))))((((((((((((........))))).))))))).......))))))))..------ ( -29.30, z-score = -3.01, R) >droYak2.chr2L 8336663 112 - 22324452 AAGUAUAGUAG-UAGAUCCAGCCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGUUUA------ ...........-...........((((.((((......((((((((.......))))))))..((((((((((((........))))).))))))).......))))))))..------ ( -21.90, z-score = -0.07, R) >droEre2.scaffold_4929 13144297 107 + 26641161 -----CAGUAG-UAUAUCCAGCCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGUUUA------ -----......-.(((((.....))))).((((.....((((((((.......))))))))..((((((((((((........))))).))))))).........))))....------ ( -22.10, z-score = -0.54, R) >droAna3.scaffold_12943 2689681 97 + 5039921 ----------------UCGCUGUGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGUUUA------ ----------------.......((((.((((......((((((((.......))))))))..((((((((((((........))))).))))))).......))))))))..------ ( -21.80, z-score = -0.98, R) >dp4.chr4_group3 9236871 109 - 11692001 CUGUCCAGCA---UCCUGCAGCCCG-GCUCGCCCCGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGCUCA------ ..(((..((.---.......))..)-)).((((.(((.((((((((.......))))))))..((((((((((((........))))).))))))).....))).))))....------ ( -25.20, z-score = 0.03, R) >droPer1.super_8 428115 109 - 3966273 CUGUCCAGCA---UCCUGCAGCCGG-GCCCGCCCCGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGCUCA------ ..((((.((.---.......)).))-)).((((.(((.((((((((.......))))))))..((((((((((((........))))).))))))).....))).))))....------ ( -29.30, z-score = -0.85, R) >droWil1.scaffold_180708 2601910 119 - 12563649 CGUUGUUCAGGCUAUCAACCACUAGCCUCCGCCCCAUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGCCGCAUUUUUUCUAC ........((((((........))))))..((..(((.((((((((.......))))))))..((((((((((((........))))).))))))).....)))..))........... ( -24.30, z-score = -1.51, R) >consensus CAGUAUAGUAG_UAUAUCCAGCCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUGACAAGUGACGAGUAAAACAAAAUGUGGCGUUUA______ .............................((((.(((.((((((((.......))))))))..((((((((((((........))))).))))))).....))).)))).......... (-19.32 = -19.33 + 0.01)

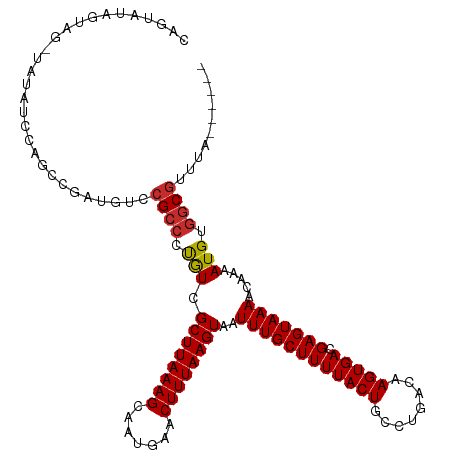
| Location | 11,916,796 – 11,916,892 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.15 |
| Shannon entropy | 0.60439 |
| G+C content | 0.48175 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -8.07 |
| Energy contribution | -8.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11916796 96 - 23011544 -------------CCCACCCCCUAUG---GAGCAGUAUUGUAGAUAUAUCCACCCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -------------.(((.......))---).((((((..(((((((((((.....)))))..........((((((((.......)))))))).))))))...))))))... ( -21.30, z-score = -3.10, R) >droSim1.chr2L 11723994 96 - 22036055 -------------CCCACCCCCUAUG---GAGCAAUAUAGUGGAUAUAUCCACCCGAUGUCCGACCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -------------............(---((((((.(((((((((((.........)))))))..)))).((((((((.......))))))))...)))))))......... ( -18.00, z-score = -1.57, R) >droSec1.super_16 116128 96 - 1878335 -------------CCCACCCCCUUUA---GAGCAAUAUAGUGGAUAUAUCCACCCGAUGUCCGCCCUGUUGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -------------.............---((((((((..((((((((.........))))))))..))))))))(((((((...((.....))...)))))))......... ( -24.10, z-score = -4.63, R) >droYak2.chr2L 8336696 98 - 22324452 -------------CCCAACCCCCAUGGUAGAGAAGUAUAGUAG-UAGAUCCAGCCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -------------............((((((((((((......-..(((...((.(....).))...)))((((((((.......))))))))....))))))).))))).. ( -21.70, z-score = -2.45, R) >droEre2.scaffold_4929 13144330 90 + 26641161 -------------CCCAACCCCCAUG--------GAGCAGUAG-UAUAUCCAGCCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -------------.(((.......))--------).(((((((-.(((((.....))))).)((...((..(((((((.......)))))))..))..))...))))))... ( -20.10, z-score = -2.50, R) >droAna3.scaffold_12943 2689714 76 + 5039921 -----------------------------------CCCAGCCCCUGU-UCGCUGUGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -----------------------------------..((((......-..))))........((...((..(((((((.......)))))))..))..))............ ( -12.70, z-score = -1.33, R) >dp4.chr4_group3 9236904 93 - 11692001 -----------------CCCCUCUCCUGCAUGUGCUGUCCAG--CAUCCUGCAGCCCGGCUCGCCCCGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -----------------........(((((.(((((....))--)))..)))))...(((..((......))..(((((((...((.....))...)))))))....))).. ( -22.20, z-score = -2.12, R) >droPer1.super_8 428148 93 - 3966273 -----------------CCCCUCUCCUGCAUGUGCUGUCCAG--CAUCCUGCAGCCGGGCCCGCCCCGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG -----------------........(((((.(((((....))--)))..))))).((((.....))))..((..(((((((...((.....))...)))))))....))... ( -24.40, z-score = -2.06, R) >droWil1.scaffold_180708 2601949 112 - 12563649 CUCACCCACAACCACCGUCUUACCCCACGAAACGUUGUUCAGGCUAUCAACCACUAGCCUCCGCCCCAUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG .......(((((...(((........)))....)))))..((((((........))))))..........((..(((((((...((.....))...)))))))....))... ( -18.60, z-score = -2.36, R) >consensus _____________CCCACCCCCCAUG___GAGCAGUAUAGUGG_UAUAUCCAACCGAUGUCCGCCCUGUCGCUUAAAGCAAUGAACUUUAAGUAAUUUGCUUUUACUGCCUG ......................................................................((..(((((((...((.....))...)))))))....))... ( -8.07 = -8.07 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:52 2011