| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,690,925 – 18,691,025 |
| Length | 100 |
| Max. P | 0.936660 |

| Location | 18,690,925 – 18,691,025 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 45.76 |
| Shannon entropy | 0.97225 |
| G+C content | 0.42270 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -5.50 |
| Energy contribution | -7.10 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18690925 100 - 22422827 ACCCACGAAAUAAUGGCCAGUAAUGGCCAAUAGUUGAA-UAGUCGUGUGCAGCCAUACAGCACAAUACAUGAC--AAUAUCAUU-CGGGCACAUUCAAAUGCAC----- .(((.(((.....((((((....))))))....)))..-..((((((((..((......))....))))))))--.........-.)))...............----- ( -27.10, z-score = -2.27, R) >droSim1.chrX 14430489 102 - 17042790 ACCCACGAAAUAAUGGCCAGUAAUGGCCAAUAGUUGAA-UAGUCGUGUGCAGCCAUACAGCACAAUACACGACCAAAUAUCAUU-CGGGCCCAUUCAAAUGCAC----- .(((.(((.....((((((....))))))....)))..-..((((((((..((......))....))))))))...........-.)))...............----- ( -28.00, z-score = -2.69, R) >droSec1.super_8 989692 82 - 3762037 ACCCACGAAAUAAUGGCCAGUAAUGGCCAAUAGUUGAA-UAGUCUUGUGCAGCCAUACAGCACAAUUCAUCAUUCAAAUGCAC-------------------------- .............((((((....))))))....(((((-(.(..((((((.........))))))..)...))))))......-------------------------- ( -21.30, z-score = -2.69, R) >droAna3.scaffold_13047 1396518 99 + 1816235 --CUUGAAGAUAUUAGCU--UUAGAGCAAGUAUUUAAAUUCGCAGUGUUCUAUUCAAAGAAGAUUCAGACCAUUUCUUCCACUU-CAAGCUGAUUAAAAUUUUG----- --(((((((.........--.(((((((.((..........))..)))))))......(((((...........)))))..)))-))))...............----- ( -16.40, z-score = -0.29, R) >droPer1.super_39 661926 105 + 745454 ----ACAGAAAUCUAGGUCCCCCCUGGAAUCGUCAAAACUCUCUUUAGGCGGGCACGGCACUCUAAUCGGAGCUGAUCCGAUCUGUGAGGCCACUAUGAAACCAAAAAC ----.......(((((((.(((((((((...............)))))).))).))(((.(((..((((((.....))))))....)))))).))).)).......... ( -29.36, z-score = -1.30, R) >consensus ACCCACGAAAUAAUGGCCAGUAAUGGCCAAUAGUUGAA_UAGUCGUGUGCAGCCAUACAGCACAAUACACCACU_AAUAGCAUU_CGAGCCCAUUAAAAUGCAC_____ .............((((((....))))))................(((((.........)))))............................................. ( -5.50 = -7.10 + 1.60)

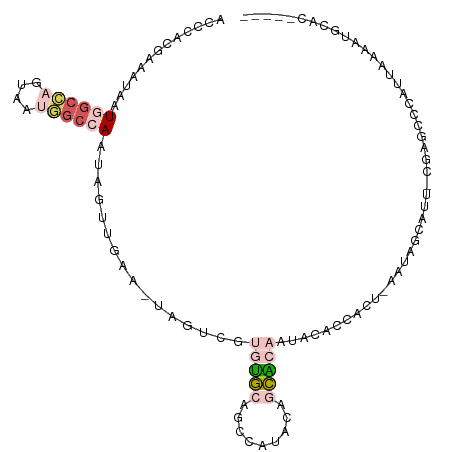
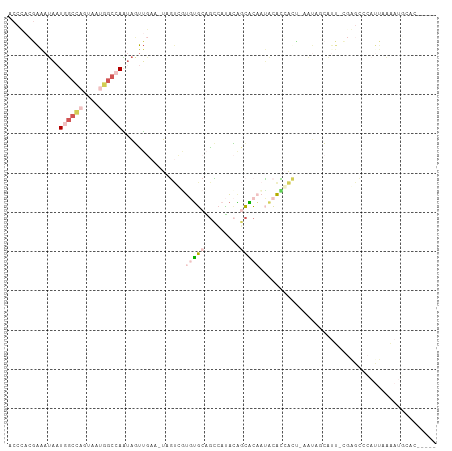
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:04 2011