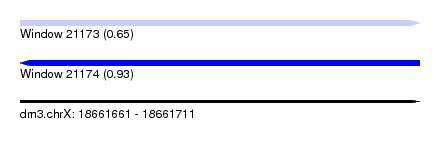
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,661,661 – 18,661,711 |
| Length | 50 |
| Max. P | 0.932434 |

| Location | 18,661,661 – 18,661,711 |
|---|---|
| Length | 50 |
| Sequences | 4 |
| Columns | 51 |
| Reading direction | forward |
| Mean pairwise identity | 79.87 |
| Shannon entropy | 0.33055 |
| G+C content | 0.54235 |
| Mean single sequence MFE | -9.10 |
| Consensus MFE | -7.85 |
| Energy contribution | -8.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.653803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
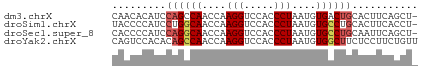
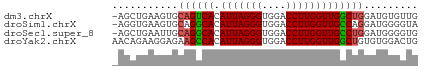
>dm3.chrX 18661661 50 + 22422827 CAACACAUCCAGCCAACCAAGGUCCACCCUAAUGUGACUGCACUUCAGCU- ..........(((.......((((((......)).))))........)))- ( -5.26, z-score = 0.38, R) >droSim1.chrX 14401210 50 + 17042790 UACCCCAUCCUGGCAACCAAGGUCCACCCUAAUGUGCCUGCACUUCACCU- ...........((((....(((.....)))....))))............- ( -8.20, z-score = -0.65, R) >droSec1.super_8 960270 50 + 3762037 CACCCCAUCCAGGCAACCAAGGUCCACCCUAAUGUGCCUGCAAUUCAGCU- .........((((((....(((.....)))....))))))..........- ( -11.90, z-score = -1.84, R) >droYak2.chrX 17283675 51 + 21770863 CAGUCCACACAGCCAACCAAGGUCCACCCUAAUGUGGCUUCUCCUUCUGUU (((.......(((((....(((.....)))....))))).......))).. ( -11.04, z-score = -1.71, R) >consensus CACCCCAUCCAGCCAACCAAGGUCCACCCUAAUGUGCCUGCACUUCAGCU_ .........((((((....(((.....)))....))))))........... ( -7.85 = -8.35 + 0.50)
| Location | 18,661,661 – 18,661,711 |
|---|---|
| Length | 50 |
| Sequences | 4 |
| Columns | 51 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Shannon entropy | 0.33055 |
| G+C content | 0.54235 |
| Mean single sequence MFE | -16.06 |
| Consensus MFE | -14.11 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
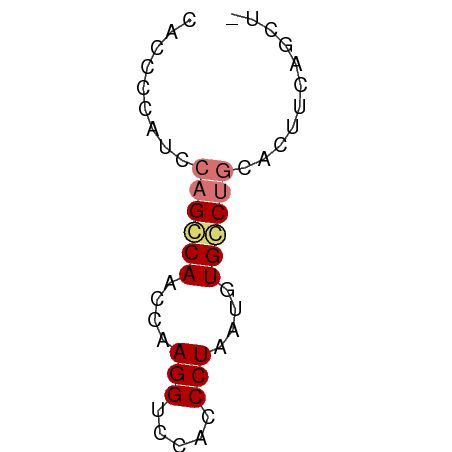
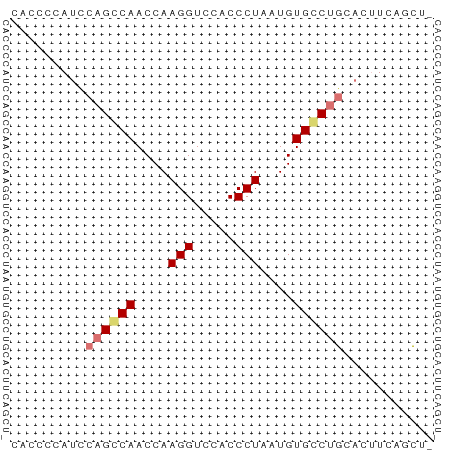
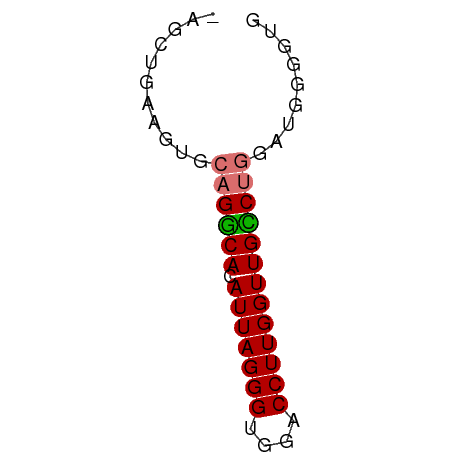
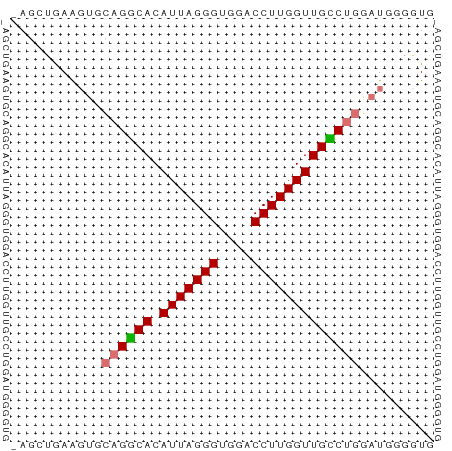
>dm3.chrX 18661661 50 - 22422827 -AGCUGAAGUGCAGUCACAUUAGGGUGGACCUUGGUUGGCUGGAUGUGUUG -(((....((.((((((.(((((((....))))))))))))).))..))). ( -16.40, z-score = -1.83, R) >droSim1.chrX 14401210 50 - 17042790 -AGGUGAAGUGCAGGCACAUUAGGGUGGACCUUGGUUGCCAGGAUGGGGUA -............((((.(((((((....)))))))))))........... ( -13.00, z-score = -0.31, R) >droSec1.super_8 960270 50 - 3762037 -AGCUGAAUUGCAGGCACAUUAGGGUGGACCUUGGUUGCCUGGAUGGGGUG -.(((..(((.((((((.(((((((....))))))))))))))))..))). ( -19.40, z-score = -2.49, R) >droYak2.chrX 17283675 51 - 21770863 AACAGAAGGAGAAGCCACAUUAGGGUGGACCUUGGUUGGCUGUGUGGACUG ..(((.......(((((.(((((((....)))))))))))).......))) ( -15.44, z-score = -1.38, R) >consensus _AGCUGAAGUGCAGGCACAUUAGGGUGGACCUUGGUUGCCUGGAUGGGGUG ...........((((((.(((((((....)))))))))))))......... (-14.11 = -14.05 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:59 2011