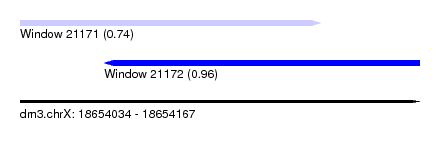
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,654,034 – 18,654,167 |
| Length | 133 |
| Max. P | 0.958972 |

| Location | 18,654,034 – 18,654,134 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.70 |
| Shannon entropy | 0.60825 |
| G+C content | 0.38586 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -8.11 |
| Energy contribution | -8.40 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
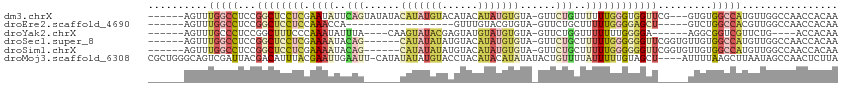
>dm3.chrX 18654034 100 + 22422827 UUCAUUUAUUCUGUUGACUCUGACUUUGUUGUGGUUGGCCAACAUGGCCA---CACCGAACCACCCAAAA-AACAGAACUACACAUAUGUAUGUACAUAUGUAU ........((((((((((.........)))((((((((((.....)))))---......)))))......-)))))))....((((((((....)))))))).. ( -27.30, z-score = -2.74, R) >droEre2.scaffold_4690 8955285 89 + 18748788 UUCAUUUAUUCUGUUGACUCUGACUUUGUUGUGGUUGGCCAACGUGGC-----CAGACAGCUCCCCAAAA-AGCAGAACUACACGUACAAACUGG--------- ........(((((((.....((.....(((((..((((((.....)))-----))))))))....))...-))))))).................--------- ( -19.70, z-score = 0.00, R) >droYak2.chrX 17276131 89 + 21770863 UUCAUUUAUUCUGUUGACUCUGACUUUGUUGUGGUCAG--AACGA--------CCGCCUUCCCCAAAAAA-ACCAGAACUACACAUACAUACUCGUAUAC---- ........(((((..(((.........)))((((((..--...))--------)))).............-..)))))......((((......))))..---- ( -13.30, z-score = -0.66, R) >droSec1.super_8 952720 100 + 3762037 UUCAUUUAUUCUGUUGACUUUGACUUUGUUGUGGUUGGCCAACAUGGCCACAACACCGAACCCCCCAAAA-AGCAGAACUACACAUAUGUACAUAUAUAUG--- ........(((((((...((((.....(((.(((((((((.....)))))....)))))))....)))).-))))))).....(((((((....)))))))--- ( -23.50, z-score = -2.17, R) >droSim1.chrX 14393642 100 + 17042790 UUCAUUUAUUCUGUUGACUCUGACUUUGUUGUGGUUGGCCAACAUGGCCACAACACCGAACCCCCCAAAA-AGCAGAACUACACAUAUGUACAUAUAUAUG--- ........(((((((.........((((.(((...(((((.....)))))..))).))))..........-))))))).....(((((((....)))))))--- ( -22.81, z-score = -1.93, R) >droMoj3.scaffold_6308 2537895 104 + 3356042 UUCAAUAACUCAAGUAAUUGUUCGCUUAUAAGAGUUGGCUAUUAAGCUUAAAAUAGCUACAAAAAUAAAACAGUAUAUAUGUAUGUAGGUACAUAUAUAUGAAU (((...(((((..((((........))))..)))))(((((((........)))))))..............((((((((((((....))))))))))))))). ( -21.40, z-score = -2.21, R) >consensus UUCAUUUAUUCUGUUGACUCUGACUUUGUUGUGGUUGGCCAACAUGGCCA___CACCGAACCCCCCAAAA_AGCAGAACUACACAUAUGUACAUAUAUAUG___ ........(((((((((((.((((...)))).))))((((.....))))......................))))))).......................... ( -8.11 = -8.40 + 0.29)

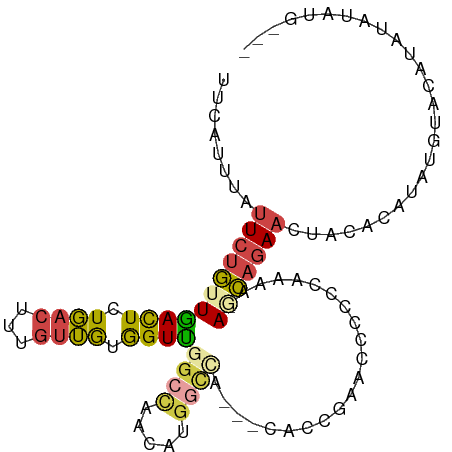
| Location | 18,654,062 – 18,654,167 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 60.62 |
| Shannon entropy | 0.71111 |
| G+C content | 0.44428 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -8.34 |
| Energy contribution | -11.02 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18654062 105 - 22422827 ------AGUUUGGCCUCCGGCUCCUCGAAUAUUCAGUAUAUACAUAUGUACAUACAUAUGUGUA-GUUCUGUUUUUUGGGUGGUUCG---GUGUGGCCAUGUUGGCCAACCACAA ------....(((((.(((((..((((((....(((..(((((((((((....)))))))))))-...)))...))))))..).)))---)...)))))(((.((....))))). ( -39.50, z-score = -3.89, R) >droEre2.scaffold_4690 8955313 85 - 18748788 ------AGUUUGGCCUCCGGCUCCUCCAAACCA------------------GUUUGUACGUGUA-GUUCUGCUUUUUGGGGAGCU-----GUCUGGCCACGUUGGCCAACCACAA ------....(((((..(((((((.(((((..(------------------((....((.....-))...))).)))))))))))-----)...))))).((.((....)))).. ( -30.40, z-score = -1.73, R) >droYak2.chrX 17276159 94 - 21770863 ------AGUUUGCCCUCCGGCUUUCCCAAAUAUUUA----CAAGUAUACGAGUAUGUAUGUGUA-GUUCUGGUUUUUUUGGGGA------AGGCGGUCGUUCUG----ACCACAA ------.(((.((...(((.((((((((((...(((----.((.((((((........))))))-.)).)))....))))))))------)).)))..))...)----))..... ( -26.70, z-score = -1.60, R) >droSec1.super_8 952748 102 - 3762037 ------AGUUUGGCCUCCGGCUCCUCGAAAAUACAG------CAUAUAUAUGUACAUAUGUGUA-GUUCUGCUUUUUGGGGGGUUCGGUGUUGUGGCCAUGUUGGCCAACCACAA ------....(((.(.(((((((((((((((((.((------(((((((((....)))))))).-))).)).))))))))))).)))).)...(((((.....))))).)))... ( -35.90, z-score = -2.18, R) >droSim1.chrX 14393670 102 - 17042790 ------AGUUUGGCCUCCGGCUCCUCGAAAAUACAG------CAUAUAUAUGUACAUAUGUGUA-GUUCUGCUUUUUGGGGGGUUCGGUGUUGUGGCCAUGUUGGCCAACCACAA ------....(((.(.(((((((((((((((((.((------(((((((((....)))))))).-))).)).))))))))))).)))).)...(((((.....))))).)))... ( -35.90, z-score = -2.18, R) >droMoj3.scaffold_6308 2537923 110 - 3356042 CGCUGGGCAGUCGAUUACGACAUUUACGAAUUGAAUU-CAUAUAUAUGUACCUACAUACAUAUAUACUGUUUUAUUUUUGUAGCU----AUUUUAAGCUUAAUAGCCAACUCUUA .....(((.((((....))))...((((((...((..-((((((((((((......)))))))))).))..))...))))))(((----......)))......)))........ ( -25.50, z-score = -3.46, R) >consensus ______AGUUUGGCCUCCGGCUCCUCCAAAAUACAG______CAUAUAUACGUACAUAUGUGUA_GUUCUGCUUUUUGGGGGGUU_____UUGUGGCCAUGUUGGCCAACCACAA .......(.((((((...((((((((.((((..(((......(((((((......)))))))......)))..)))).))))............)))).....)))))).).... ( -8.34 = -11.02 + 2.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:57 2011