| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,903,547 – 11,903,648 |
| Length | 101 |
| Max. P | 0.981136 |

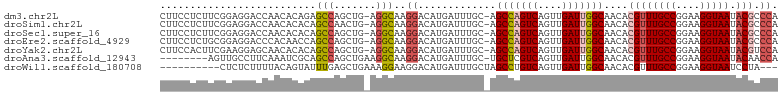
| Location | 11,903,547 – 11,903,648 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Shannon entropy | 0.36770 |
| G+C content | 0.51229 |
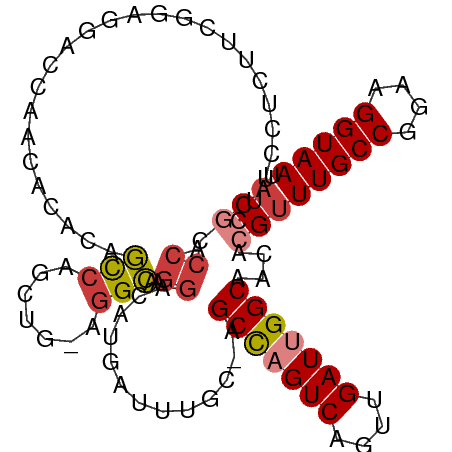
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -21.89 |
| Energy contribution | -22.54 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
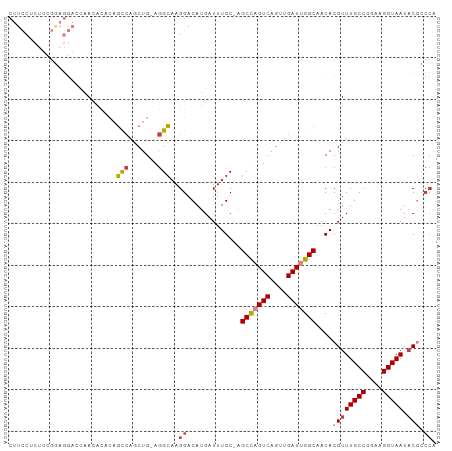
>dm3.chr2L 11903547 101 - 23011544 CUUCCUCUUCGGAGGACCAACACAGAGCCAGCUG-AGGCAAGGACAUGAUUUGC-AGCCAGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUACGCCCA ..(((((....)))))..........(((.....-.)))..((...........-.(((((((....)))))))....((((((((....))))).))).)). ( -37.50, z-score = -2.96, R) >droSim1.chr2L 11710342 101 - 22036055 CUUCCUCUUCGGAGGACCAACACACAGCCAACUG-AGGCAAGGACAUGAUUUGC-AGCCAGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUACGCCCA ..(((((....)))))..........(((.....-.)))..((...........-.(((((((....)))))))....((((((((....))))).))).)). ( -37.50, z-score = -3.47, R) >droSec1.super_16 102997 101 - 1878335 CUUCCUCUUCGGAGGACCAACACACAGCCAGCUG-AGGCAAGGACAUGAUUUGC-AGCCAGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUACGCCCA ..(((((....)))))..........(((.....-.)))..((...........-.(((((((....)))))))....((((((((....))))).))).)). ( -37.50, z-score = -3.10, R) >droEre2.scaffold_4929 13131355 101 + 26641161 CUUCCUCUGCGGAGGACCCACAACCAGCCAGCUG-AGGCAAGGACAUGAUUUGC-AGCCAGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUACGCCCA ..(((((....)))))..........(((.....-.)))..((...........-.(((((((....)))))))....((((((((....))))).))).)). ( -34.50, z-score = -1.52, R) >droYak2.chr2L 8322845 101 - 22324452 CUUCCACUUCGAAGGAGCAACACACAGCCAGCUG-AGGCAAGGACAUGAUUUGC-AGCCAGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUACGUCCA .((((........)))).........(((.....-.)))..((((.........-.(((((((....))))))).....(((((((....))))).)))))). ( -34.10, z-score = -2.54, R) >droAna3.scaffold_12943 2676369 94 + 5039921 --------AGUUGCCUUCAAAUCGCAGCCAGCUGAAGGCAAGGACAUGAUUUGC-UGCUCGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUACAACCA --------.(((((((((..........(((((((.((((((.......)))))-).....)))))))..((((((.....)))))))))))))))....... ( -32.40, z-score = -2.07, R) >droWil1.scaffold_180708 2582849 90 - 12563649 ----------CUCUCUUUUACAGUAUUUGAGCUGAAAGGAAGGACAUGAUUUGCUAGCCUGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUCCUA--- ----------...((((((.((((......))))))))))((((............(((.(((....))).))).......(((((....))))))))).--- ( -26.80, z-score = -1.85, R) >consensus CUUCCUCUUCGGAGGACCAACACACAGCCAGCUG_AGGCAAGGACAUGAUUUGC_AGCCAGUCAGUUGAUUGGCAACACGUUUGCCGGAAGGUAAUACGCCCA ..........................(((.......)))..((.............(((((((....)))))))....((((((((....))))).))).)). (-21.89 = -22.54 + 0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:49 2011