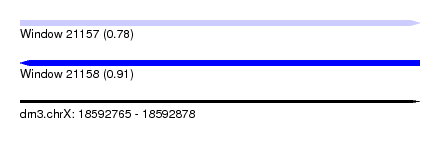
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,592,765 – 18,592,878 |
| Length | 113 |
| Max. P | 0.910246 |

| Location | 18,592,765 – 18,592,878 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.21 |
| Shannon entropy | 0.20502 |
| G+C content | 0.40722 |
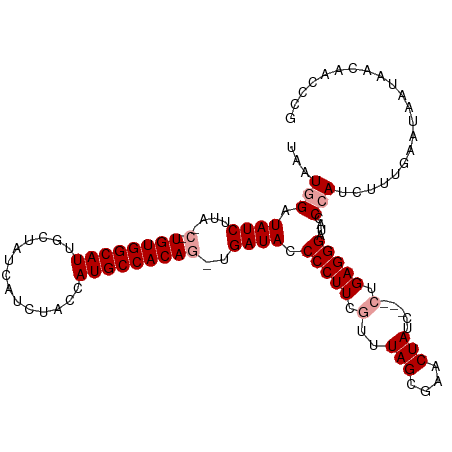
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -22.27 |
| Energy contribution | -22.61 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18592765 113 + 22422827 CGGGUUGUUAUUAUUCAAAAAUGGCUAACCCUCAGGUGGAUAGUUCGCUAAAUGAAGGGGUAUCA-CUGUGGCAUUAGAAAUGAUAGCAAUGCCACACGAUAAGAUAUCCAUUA .(((((((((((.......)))))).)))))...((((((((..((.((......)).))((((.-.(((((((((............))))))))).))))...)))))))). ( -32.50, z-score = -2.32, R) >droSec1.super_8 895457 105 + 3762037 CGGUUUGUUAUUAUUCAAAGAUGGGUAUCCCUCAG---GAUAGUUUGCUAAACGAAGGGUUAUCA-CUGUGGCAUGGUAGAUGAUAGCAAUGCCACA-----AGAUAUCCAUUA ...((((........))))(((((((((((((..(---..(((....)))..)..))).......-.((((((((.((........)).))))))))-----.)))))))))). ( -32.00, z-score = -2.62, R) >droSim1.chrX_random 4841567 111 + 5698898 CGGGUUGUUAUUAUUCAAAGAUGGGUAUCCCUCAG---GAUAGUUCACUAAGCGAAGGGGUAUCAACUGUGGCAUGGUAGAUGAUAGCAAUGCCACAAGGUAAGAUAUCUAGUA .((((.......))))..((((((((((((((..(---..(((....)))..)..)))))))))...((((((((.((........)).))))))))........))))).... ( -31.20, z-score = -1.83, R) >consensus CGGGUUGUUAUUAUUCAAAGAUGGGUAUCCCUCAG___GAUAGUUCGCUAAACGAAGGGGUAUCA_CUGUGGCAUGGUAGAUGAUAGCAAUGCCACA_G_UAAGAUAUCCAUUA ((....((.((((((.......((.....)).......))))))..))....))...(((((((...((((((((.((........)).))))))))......))))))).... (-22.27 = -22.61 + 0.34)

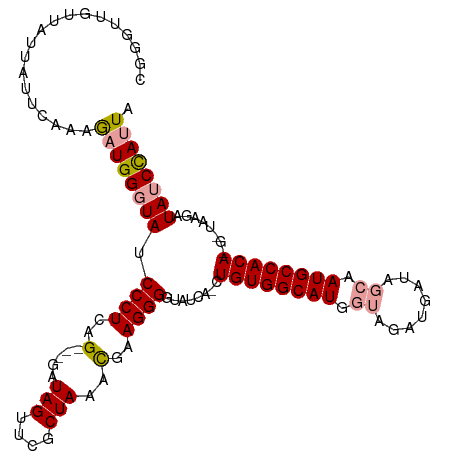
| Location | 18,592,765 – 18,592,878 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.21 |
| Shannon entropy | 0.20502 |
| G+C content | 0.40722 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -17.67 |
| Energy contribution | -18.01 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18592765 113 - 22422827 UAAUGGAUAUCUUAUCGUGUGGCAUUGCUAUCAUUUCUAAUGCCACAG-UGAUACCCCUUCAUUUAGCGAACUAUCCACCUGAGGGUUAGCCAUUUUUGAAUAAUAACAACCCG ...((((((.....(((((((((((((..(....)..)))))))))((-(((.......)))))..))))..)))))).....(((((.(..(((.......)))..)))))). ( -26.80, z-score = -2.24, R) >droSec1.super_8 895457 105 - 3762037 UAAUGGAUAUCU-----UGUGGCAUUGCUAUCAUCUACCAUGCCACAG-UGAUAACCCUUCGUUUAGCAAACUAUC---CUGAGGGAUACCCAUCUUUGAAUAAUAACAAACCG ..((((.(((((-----((((((((.(..........).)))))))))-.)))).(((((.(..(((....)))..---).)))))....)))).................... ( -26.90, z-score = -3.27, R) >droSim1.chrX_random 4841567 111 - 5698898 UACUAGAUAUCUUACCUUGUGGCAUUGCUAUCAUCUACCAUGCCACAGUUGAUACCCCUUCGCUUAGUGAACUAUC---CUGAGGGAUACCCAUCUUUGAAUAAUAACAACCCG .................((((((((.(..........).))))))))((((....(((((.(..(((....)))..---).))))).....((....))........))))... ( -23.20, z-score = -2.28, R) >consensus UAAUGGAUAUCUUA_C_UGUGGCAUUGCUAUCAUCUACCAUGCCACAG_UGAUACCCCUUCGUUUAGCGAACUAUC___CUGAGGGAUACCCAUCUUUGAAUAAUAACAACCCG ...(((.((((......((((((((..............))))))))...)))).(((((....(((....))).......)))))....)))..................... (-17.67 = -18.01 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:45 2011