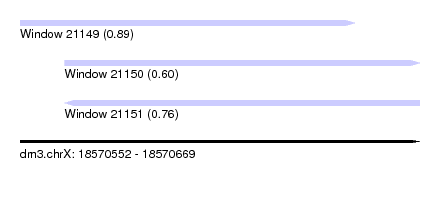
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,570,552 – 18,570,669 |
| Length | 117 |
| Max. P | 0.887964 |

| Location | 18,570,552 – 18,570,650 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.68 |
| Shannon entropy | 0.35287 |
| G+C content | 0.58574 |
| Mean single sequence MFE | -41.39 |
| Consensus MFE | -31.58 |
| Energy contribution | -32.01 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18570552 98 + 22422827 -------AGCAAUGCUGUUUUUUAGUCGGUCGGUCAUUGACCGUUGC--GCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGCU-GCGGCAACU--------- -------.(((..(((((.((((((((((.(((((...)))))))).--))))))).)))))...((((((((..--(((((...))))).)))))))))-))(....).--------- ( -43.30, z-score = -1.59, R) >droSim1.chrX 14331090 98 + 17042790 -------AGCAAUGCUGUUUUUUAGUCGGUCGGUCAUUGACCGUUGC--GCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGCU-GCGGCAACU--------- -------.(((..(((((.((((((((((.(((((...)))))))).--))))))).)))))...((((((((..--(((((...))))).)))))))))-))(....).--------- ( -43.30, z-score = -1.59, R) >droSec1.super_8 874248 98 + 3762037 -------AGCAAUGCUGUUUUUUAGUCGGUCGGUCAUUGACCGUUGC--GCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGUU-GCGGCAACU--------- -------.((((((((((.((((((((((.(((((...)))))))).--))))))).)))))..((((((.((((--(....)))))....)))))))))-))(....).--------- ( -42.00, z-score = -1.46, R) >droYak2.chrX 17196994 98 + 21770863 -------AGCAAUGCUGUUUUUUAGUCGGUCGGUCAUUGACCGUUGC--GCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGCU-GCGGCAACU--------- -------.(((..(((((.((((((((((.(((((...)))))))).--))))))).)))))...((((((((..--(((((...))))).)))))))))-))(....).--------- ( -43.30, z-score = -1.59, R) >droEre2.scaffold_4690 8879910 90 + 18748788 -------AGCAAUGCUGUUUUU-AGUCGGUCGGUCAUUGACCGUUGC--GCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGCU-GC---------------- -------.(((..(((((((((-((((((.(((((...)))))))).--))))))).)))))...((((((((..--(((((...))))).)))))))))-))---------------- ( -39.30, z-score = -1.45, R) >droAna3.scaffold_13248 1742338 104 + 4840945 GUGCGACAGCAAUGCUGUUUUUUAGUCGGUCGGUCAUUGACCGUUUC----UGAGACGCAGUGACGCCGUGGCUGGCUUUUCGAAGGGGGUACGGUAGU--GCGGUGGUG--------- .(((....)))..(((((.((((((.((((((.....))))))...)----))))).)))))..((((((.((((...(..(....)..)..))))...--))))))...--------- ( -39.30, z-score = -1.61, R) >droWil1.scaffold_181096 11206143 105 + 12416693 ------------UUUUGUUUUUUAGUCGGUCGGUCAUUGACCGUUUACAACUGAGACGCAUUGUCGCCGCUGCCA--UUGGCAUGGUCGACGCUGUGGCUUGCAGUUGCCGCUGCUGUU ------------...(((.(((((((.(..(((((...)))))....).))))))).)))..(((((((.((((.--..))))))).))))((.(((((........))))).)).... ( -39.20, z-score = -2.07, R) >consensus _______AGCAAUGCUGUUUUUUAGUCGGUCGGUCAUUGACCGUUGC__GCUGAGACGCAGUGACGCCGCUGCCA__UUGGCAUGGCCGACGCGGUGGCU_GCGGCAACU_________ ........((...(((((.(((((((....(((((...)))))......))))))).)))))...((((((((....(((((...))))).))))))))..))................ (-31.58 = -32.01 + 0.43)

| Location | 18,570,565 – 18,570,669 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.09 |
| Shannon entropy | 0.42647 |
| G+C content | 0.62219 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.02 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18570565 104 + 22422827 UUUAGUCGGUCGGUCAUUGACCGUUGCGCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGCUGCGGCAACUGAGGUAGCUGAGGCAACUG ....(((((((((((...)))).....((..(...((((((....))))))..--)..))..))))))).((((.(((.((((.((....)).)))).))).)))) ( -45.30, z-score = -1.22, R) >droSim1.chrX 14331103 95 + 17042790 UUUAGUCGGUCGGUCAUUGACCGUUGCGCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGCUGCGGCAACUGAGGCAGCUG--------- ....(((((((((((...)))).....((..(...((((((....))))))..--)..))..))))))).....((((((..(....)..)))))).--------- ( -41.80, z-score = -0.87, R) >droEre2.scaffold_4690 8879922 78 + 18748788 UUUAGUCGGUCGGUCAUUGACCGUUGCGCUGAGACGCAGUGACGCCGCUGCCA--UUGGCAUGGCCGACGCGGUGGCUGC-------------------------- ....((((((((.....))))).(((((......))))).)))((((((((..--(((((...))))).))))))))...-------------------------- ( -33.50, z-score = -0.95, R) >droAna3.scaffold_13248 1742358 93 + 4840945 UUUAGUCGGUCGGUCAUUGACCGUUUC--UGAGACGCAGUGACGCCGUGGCUGGCUUUUCGAAGGGGGUACGGUAG-UGCGGUGGUGGGGUUAGUG---------- .....((((.(((((...)))))...)--)))(((.(.....((((((.((((...(..(....)..)..))))..-.))))))...).)))....---------- ( -30.20, z-score = -0.59, R) >consensus UUUAGUCGGUCGGUCAUUGACCGUUGCGCUGAGACGCAGUGACGCCGCUGCCA__UUGGCAUGGCCGACGCGGUGGCUGCGGCAACUGAGGUAGCU__________ ....((((((((.....))))).(((((......))))).)))(((((.((((..(((((...))))).....)))).)))))....................... (-27.34 = -27.02 + -0.31)



| Location | 18,570,565 – 18,570,669 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.09 |
| Shannon entropy | 0.42647 |
| G+C content | 0.62219 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18570565 104 - 22422827 CAGUUGCCUCAGCUACCUCAGUUGCCGCAGCCACCGCGUCGGCCAUGCCAA--UGGCAGCGGCGUCACUGCGUCUCAGCGCAACGGUCAAUGACCGACCGACUAAA ..(((((..(((((.....)))))..)))))..((((....(((((....)--)))).)))).(((..(((((....))))).(((((...)))))...))).... ( -39.90, z-score = -1.84, R) >droSim1.chrX 14331103 95 - 17042790 ---------CAGCUGCCUCAGUUGCCGCAGCCACCGCGUCGGCCAUGCCAA--UGGCAGCGGCGUCACUGCGUCUCAGCGCAACGGUCAAUGACCGACCGACUAAA ---------.(((((...)))))(((((.((((..((((.....))))...--)))).)))))(((..(((((....))))).(((((...)))))...))).... ( -36.80, z-score = -1.13, R) >droEre2.scaffold_4690 8879922 78 - 18748788 --------------------------GCAGCCACCGCGUCGGCCAUGCCAA--UGGCAGCGGCGUCACUGCGUCUCAGCGCAACGGUCAAUGACCGACCGACUAAA --------------------------.......((((....(((((....)--)))).)))).(((..(((((....))))).(((((...)))))...))).... ( -29.70, z-score = -1.22, R) >droAna3.scaffold_13248 1742358 93 - 4840945 ----------CACUAACCCCACCACCGCA-CUACCGUACCCCCUUCGAAAAGCCAGCCACGGCGUCACUGCGUCUCA--GAAACGGUCAAUGACCGACCGACUAAA ----------...............((((-(....)).........((...(((......))).))...))).....--....(((((.......)))))...... ( -15.80, z-score = -0.37, R) >consensus __________AGCUACCUCAGUUGCCGCAGCCACCGCGUCGGCCAUGCCAA__UGGCAGCGGCGUCACUGCGUCUCAGCGCAACGGUCAAUGACCGACCGACUAAA .........................(((((.(.((((....((((........)))).)))).)...)))))...........(((((.......)))))...... (-22.06 = -22.12 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:40 2011