
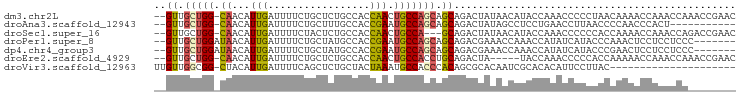
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,892,943 – 11,893,037 |
| Length | 94 |
| Max. P | 0.839922 |

| Location | 11,892,943 – 11,893,037 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 66.17 |
| Shannon entropy | 0.65354 |
| G+C content | 0.48542 |
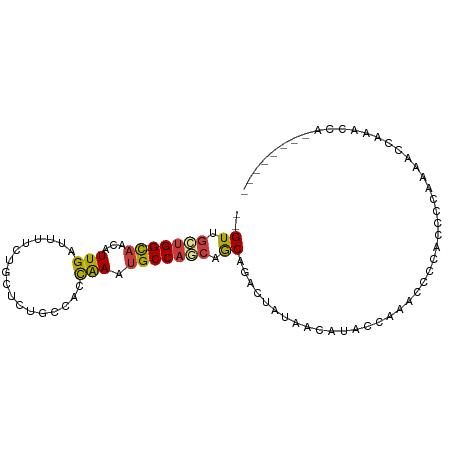
| Mean single sequence MFE | -13.35 |
| Consensus MFE | -5.53 |
| Energy contribution | -5.39 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839922 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 11892943 94 - 23011544 --GUUGCUGG-CAACAUUGAUUUUCUGCUCUGCCACCAACUGCCAGCAGCAGACUAUAACAUACCAAACCCCCUAACAAAACCAAACCAAACCGAAC --((((((((-((...(((.................))).))))))))))............................................... ( -16.63, z-score = -2.90, R) >droAna3.scaffold_12943 2668138 83 + 5039921 --GUUGCUGG-CAACAUUGAUUUUCUGCUUUGCCACCGAAUGCCAGCAGCAGACUAUAGCCUCCUGAACCUUAACCCCAACCCACU----------- --((((((((-((...(((.................))).))))))))))....................................----------- ( -16.33, z-score = -1.59, R) >droSec1.super_16 95694 91 - 1878335 --GUUGCUGG-CAACAUUGAUUUUCUACUCUGCCACCAACUGCCA---GCAGACUAUAACAUACCAAACCCCCCACCAAAACCAAACCAGACCGAAC --.(((((((-((...(((.................))).)))))---))))............................................. ( -12.93, z-score = -2.48, R) >droPer1.super_8 406233 88 - 3966273 --GUUGCUGGAUAACAUUGAUUUUCUGCUAUGCCACCGAAUGCCAGCAGCAGACGAAACCAAACCAUAUCAUACCCAAACUCCUCCUCCC------- --((((((((.......((..............)).......))))))))........................................------- ( -12.08, z-score = -0.80, R) >dp4.chr4_group3 9217743 88 - 11692001 --GUUGCUGGAUAACAUUGAUUUUCUGCUAUGCCACCGAAUGCCAGCAGCAGACGAAACCAAACCAUAUCAUACCCGAACUCCUCCUCCC------- --((((((((.......((..............)).......))))))))........................................------- ( -12.08, z-score = -0.55, R) >droEre2.scaffold_4929 13123847 89 + 26641161 --GUUGCUGG-CAACAUUGAUUUUCUGCUCUGCCACCAACUGCCACCUGCAGACUA-----UACCAAACCCCCACCAAAAACCAAACCAAACCGAAC --.(((.(((-((.....(.....).....))))).)))((((.....))))....-----.................................... ( -11.40, z-score = -1.62, R) >droVir3.scaffold_12963 6914889 75 + 20206255 UUGUUGGCGG-CUACAUUGAUUUUCAGCUCUGCUACUAAAUGCCACCCACAGCGCACAAUCGCACACAUUCCUUAC--------------------- .(((.((.((-(......(.....)(((...))).......))).)).)))(((......))).............--------------------- ( -12.00, z-score = 0.06, R) >consensus __GUUGCUGG_CAACAUUGAUUUUCUGCUCUGCCACCAAAUGCCAGCAGCAGACUAUAACAUACCAAACCCCACCCCAAAACCAAACCA________ ..((.(((((.((...(((.................))).))))))).))............................................... ( -5.53 = -5.39 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:48 2011