
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,546,053 – 18,546,149 |
| Length | 96 |
| Max. P | 0.606409 |

| Location | 18,546,053 – 18,546,149 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 57.09 |
| Shannon entropy | 0.82499 |
| G+C content | 0.62085 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -12.19 |
| Energy contribution | -12.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
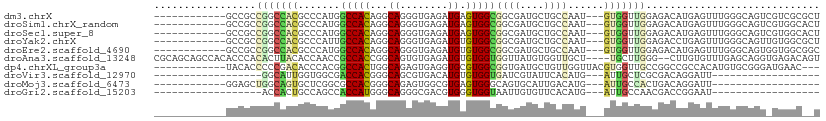
>dm3.chrX 18546053 96 + 22422827 ------------GCCGCCGGCCACGCCCAUGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAU---GUGGUUGGAGACAUGAGUUUGGGCAGUCGUCGCGCU ------------(((((((..(.(((((...(((...))).))))).).)).))))).(((((((((((...---.....((....))........))))).))))))... ( -44.49, z-score = -1.36, R) >droSim1.chrX_random 4810929 96 + 5698898 ------------GCCGCCGGCCACGCCCAUGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAU---GUGGUUGGAGACAUGAGUUUGGGCAGUCGUGGCACU ------------((((((((((((((((...(((...))).))))........(((((((...)))))))..---)))))))..(((.((........))))))))))... ( -41.60, z-score = -0.80, R) >droSec1.super_8 849513 96 + 3762037 ------------GCCGCCGGCCACGCCCAUGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAU---GUGGUUGGAGACAUGAGUUUGGGCAGUCGUGGCACU ------------((((((((((((((((...(((...))).))))........(((((((...)))))))..---)))))))..(((.((........))))))))))... ( -41.60, z-score = -0.80, R) >droYak2.chrX 17171804 96 + 21770863 ------------GCCGCCGGCCACGCCCAUUGCCACAGGCAGGGUGAGAUGUGUGGCGGCGAUGCUGCCAAU---GUGGUUGGAGACCUGAGUUUGGGCAGUUGUGGCGCU ------------((((((((((((((((..((((...))))))))........(((((((...)))))))..---)))))))..(.(((......))))....)))))... ( -41.30, z-score = -0.33, R) >droEre2.scaffold_4690 8855740 96 + 18748788 ------------GCCGCCGGCCACGCCCAUGGCCACAGGCAGGGUGAGAUGUGUGGCGGCGAUGCUGCCAAU---GUGGUUGGAGACAUGAGUUUGGGCAGUGGUGGCGGC ------------(((((((.(((((((((.(((((((.(((........))).(((((((...))))))).)---))))))(....).......))))).))))))))))) ( -51.70, z-score = -3.45, R) >droAna3.scaffold_13248 1714099 105 + 4840945 CGCAGCAGCCACACCCACACUUACACCAACCGCCACCGGCAGUGUGAGAUGUGUGGUGGUUAUGUGGUUGCU----UGCUUGGG--CUUGUGUUUGAGCAGGUGAGACAGU ((((..((((((.(.(((((((((((...(((....)))..))))))).)))).))))))).)))).(..((----((((..((--(....)))..))))))..)...... ( -46.10, z-score = -2.28, R) >dp4.chrXL_group3a 2004588 96 - 2690836 ------------UACACCCCCGACACCCACGGCCACUGGCAGAGUGAGGUGCGUGGCGGUGAUGCUGUUGGUUACGUGGUUGCCGGCCGCCACAUGUGCGGGAUGAAC--- ------------......((((.(((((((.(((...)))...))).)))).((((((((........((((.((...)).)))))))))))).....))))......--- ( -37.40, z-score = 0.40, R) >droVir3.scaffold_12970 5357918 72 + 11907090 ------------------GGCAUUGGUGGCGACCACGGGCAGCGUGACAUGUGUGGUGAUCGUAUUCACAUG---AUUGCUCGCGACAGGAUU------------------ ------------------.......((.((((.((((.....)))).((((((.((((....))))))))))---.....)))).))......------------------ ( -19.00, z-score = 1.29, R) >droMoj3.scaffold_6473 4940148 78 + 16943266 ------------GGAGCUGGCAGUGCUCGGCGCCACGGGCAGAGUGGCGUGAGUGGGCAGUGCAUUGACAUG---AUUGCCACUGACAGGAUU------------------ ------------....(((.(((((((((.((((((.......))))))))))).((((((.(((....)))---)))))))))).)))....------------------ ( -33.60, z-score = -2.11, R) >droGri2.scaffold_15203 2518162 72 - 11997470 ------------------ACCACUGCCAGCCACCAUGGGCAGGGCGACGUGGGUGGUAAUUGUGUUCACAUG---AUUGCCAACGACCGGAAU------------------ ------------------.((((((((.(((......)))..))))..)))).(((((((..((....))..---)))))))...........------------------ ( -27.60, z-score = -1.25, R) >consensus ____________GCCGCCGGCCACGCCCAUGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAU___GUGGUUGGAGACAUGAGUUUGGGCAGU_GUGGC__U .................(((((((.......(((((...((........)).)))))((((....))))......)))))))............................. (-12.19 = -12.52 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:35 2011