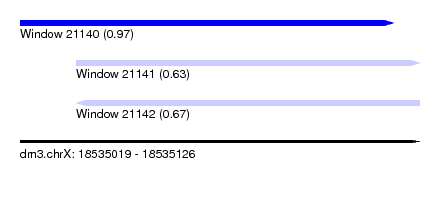
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,535,019 – 18,535,126 |
| Length | 107 |
| Max. P | 0.974313 |

| Location | 18,535,019 – 18,535,119 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Shannon entropy | 0.32270 |
| G+C content | 0.48248 |
| Mean single sequence MFE | -37.14 |
| Consensus MFE | -26.61 |
| Energy contribution | -28.29 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18535019 100 + 22422827 ACGGAGCAUCGGCAACAU-UAUUU-GGCUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUCAGAUU-------------GGUUGUCUGUGGUGUUUCUUUUCUGUUAUU ..((((((((((((((..-.((((-((((((((((((((((....)))))).....))))))))).))))).-------------.))))))...))))))))............ ( -39.50, z-score = -4.18, R) >droSim1.chrX 14305943 113 + 17042790 ACGGAGCAUCGGCAACAU-UAUUU-GGCUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUCAGAUUCGCAUUUUGCGUUGGUUGCCUGUGGUGUUUGUUUUCUGCUCCU ..((((((..((((((..-.((((-((((((((((((((((....)))))).....))))))))).))))).(((.....)))...))))))...((.........)))))))). ( -43.10, z-score = -2.72, R) >droSec1.super_8 838766 113 + 3762037 ACGGAGCAUCGGCAACAU-UAUUU-AGCUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUCAGAUUCGCAUUUUGCGUUGGUUGCCUGUGGUGUUUGUUUUCUGCUCCU ..((((((..((((((..-.((((-((((((((((((((((....)))))).....)))))))))).)))).(((.....)))...))))))...((.........)))))))). ( -41.10, z-score = -2.30, R) >droYak2.chrX 17161137 103 + 21770863 ACGGAGCAGCGGCAUCAU-UUUUU-GACUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUUAGAUUC---------GCAUUUUGCGC-UGGUUUUUCUACUCGGUUUUU ((.(((.((((((.....-..(((-((((((((((((((((....)))))).....)))))))).)))))...---------((.....))))-))......)).))).)).... ( -27.40, z-score = -0.17, R) >droEre2.scaffold_4690 8844592 105 + 18748788 ACGGAGCAUCGGCAACAUAUAUUUUGGCUAGUGAUUCACUCGAGCGAUUGACGACAAUCGCUGGCUUGGAUU----------GCCUGUGGUGUUGGGUGUUUCUUUUCUGGUUUU ..((((((((..((((((.(((...(((....((((((..(.((((((((....)))))))).)..))))))----------))).)))))))))))))))))............ ( -34.60, z-score = -2.46, R) >consensus ACGGAGCAUCGGCAACAU_UAUUU_GGCUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUCAGAUUC_________GCUGGUUGCCUGUGGUGUUUCUUUUCUGCUCUU ..((((((((((((((....((((.((((((((((((((((....)))))).....)))))))))).))))...............))))))...))))))))............ (-26.61 = -28.29 + 1.68)


| Location | 18,535,034 – 18,535,126 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.57 |
| Shannon entropy | 0.36703 |
| G+C content | 0.42347 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18535034 92 + 22422827 --CAUUAUUUGGCUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUCAGAUU-------------GGUUGUCUGUGGUGUUUCUUUUCUGUUAUUAUUUUUU --(((((...((((((((((((((((....)))))).....))))))))))(((((.-------------....))))))))))....................... ( -24.80, z-score = -1.92, R) >droSec1.super_8 838781 105 + 3762037 --CAUUAUUUAGCUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUCAGAUUCGCAUUUUGCGUUGGUUGCCUGUGGUGUUUGUUUUCUGCUCCUAUUUUUU --........((((((((((((((((....)))))).....))))))))))((((((((((....(((.....))).))))).)))))................... ( -26.20, z-score = -0.08, R) >droYak2.chrX 17161152 95 + 21770863 --CAUUUUUUGACUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUUAGAUUCGC----------AUUUUGCGCUGGUUUUUCUACUCGGUUUUUAUUUUUU --.....(((((((((((((((((((....)))))).....)))))))).)))))...((----------.....))((((((......)).))))........... ( -21.80, z-score = -0.90, R) >droEre2.scaffold_4690 8844607 97 + 18748788 CAUAUAUUUUGGCUAGUGAUUCACUCGAGCGAUUGACGACAAUCGCUGGCUUGGAUUGCC----------UGUGGUGUUGGGUGUUUCUUUUCUGGUUUUAUUUCUU ..........((((((.((..((((((((((((((....))))))).(((.......)))----------.......)))))))..))....))))))......... ( -28.20, z-score = -2.70, R) >consensus __CAUUAUUUGGCUAGUGAUUCACUCGAGCGAGUGGCGACAAUCGCUGGCUCAGAUUCGC__________GGUUGUCUGUGGUGUUUCUUUUCUGUUUUUAUUUUUU ..........((((((((((((((((....)))))).....))))))))))........................................................ (-17.25 = -17.38 + 0.12)

| Location | 18,535,034 – 18,535,126 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Shannon entropy | 0.36703 |
| G+C content | 0.42347 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -9.36 |
| Energy contribution | -9.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18535034 92 - 22422827 AAAAAAUAAUAACAGAAAAGAAACACCACAGACAACC-------------AAUCUGAGCCAGCGAUUGUCGCCACUCGCUCGAGUGAAUCACUAGCCAAAUAAUG-- ............................((((.....-------------..)))).((.((.((((.....(((((....))))))))).)).)).........-- ( -13.30, z-score = -1.52, R) >droSec1.super_8 838781 105 - 3762037 AAAAAAUAGGAGCAGAAAACAAACACCACAGGCAACCAACGCAAAAUGCGAAUCUGAGCCAGCGAUUGUCGCCACUCGCUCGAGUGAAUCACUAGCUAAAUAAUG-- ........((..((((..............(....)...(((.....)))..))))..)).(((.....)))(((((....)))))...................-- ( -22.90, z-score = -1.66, R) >droYak2.chrX 17161152 95 - 21770863 AAAAAAUAAAAACCGAGUAGAAAAACCAGCGCAAAAU----------GCGAAUCUAAGCCAGCGAUUGUCGCCACUCGCUCGAGUGAAUCACUAGUCAAAAAAUG-- ..............((.(((.........(((.....----------)))...........(((.....)))(((((....))))).....))).))........-- ( -18.00, z-score = -1.78, R) >droEre2.scaffold_4690 8844607 97 - 18748788 AAGAAAUAAAACCAGAAAAGAAACACCCAACACCACA----------GGCAAUCCAAGCCAGCGAUUGUCGUCAAUCGCUCGAGUGAAUCACUAGCCAAAAUAUAUG ...................((..(((.(.........----------(((.......)))((((((((....)))))))).).)))..))................. ( -18.00, z-score = -3.22, R) >consensus AAAAAAUAAAAACAGAAAAGAAACACCACAGACAACC__________GCGAAUCUAAGCCAGCGAUUGUCGCCACUCGCUCGAGUGAAUCACUAGCCAAAAAAUG__ .........................................................((.((.((((.....(((((....))))))))).)).))........... ( -9.36 = -9.42 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:33 2011