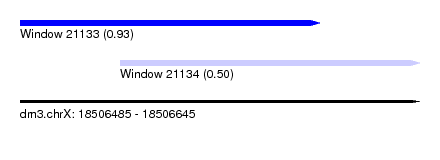
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,506,485 – 18,506,645 |
| Length | 160 |
| Max. P | 0.933297 |

| Location | 18,506,485 – 18,506,605 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Shannon entropy | 0.17947 |
| G+C content | 0.59444 |
| Mean single sequence MFE | -54.47 |
| Consensus MFE | -43.98 |
| Energy contribution | -45.10 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18506485 120 + 22422827 UAAGGGCCAGGGCAGUGGAGGCCAAGGACAUGCUGGAUGUGUUGGACAUUUCCGCAUUCGGUAUGGGGCUGACCAAGUCUGUGGAGCAUAGCUCCUCGCCGGGCACUGCCGGUUCCGUCA ...(((((..((((((((.((((.....((((((((((((...(((....))))))))))))))).))))..))..(((((.(((((...)))))....))))))))))))))))..... ( -52.80, z-score = -1.72, R) >droYak2.chrX 17134556 120 + 21770863 UGAAAUCCAGGGCAGUGGUGGCCAAGGACAUGCUGGAUGUGCUGGAGAUGUCCGCAUUCGGCAUGGAGCUGACCCAGUCUGAGGAGCACAGCUCCUCGCUGGGCACCACCGGGUCCGACA .........((((..((((((.......((((((((((((...(((....)))))))))))))))....((.((((((..(((((((...))))))))))))))))))))).)))).... ( -57.10, z-score = -2.31, R) >droSec1.super_8 810670 120 + 3762037 UAAGUGCCAGGGCAGUGGAAGCCAAGGACAUGCUGGAUGUGUUGGAAUUUUCACCAUUCGGUAUGGGGCUGACCAAGUCUGAGGAGCAUAGCUCCUCGCCGGGCACUGCCGGUUCCGUCA ..((((((..(((..(((.((((.....(((((((((((.((.(((...)))))))))))))))).))))..))).....(((((((...)))))))))).))))))(.((....)).). ( -53.50, z-score = -2.91, R) >consensus UAAGAGCCAGGGCAGUGGAGGCCAAGGACAUGCUGGAUGUGUUGGAAAUUUCCGCAUUCGGUAUGGGGCUGACCAAGUCUGAGGAGCAUAGCUCCUCGCCGGGCACUGCCGGUUCCGUCA ...(((((..((((((....(((.....((((((((((((...(((....))))))))))))))).(((.(((...))).(((((((...)))))))))).))))))))))))))..... (-43.98 = -45.10 + 1.12)

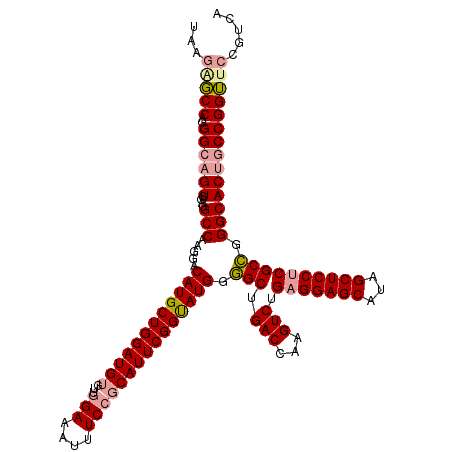
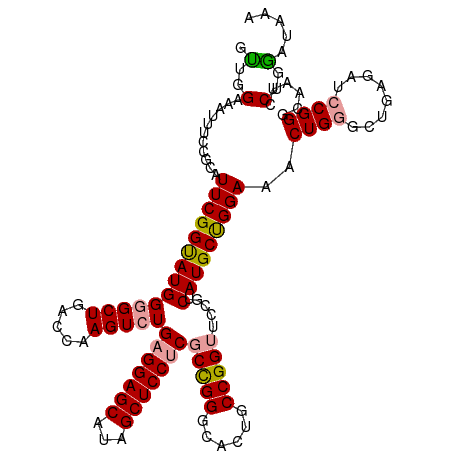
| Location | 18,506,525 – 18,506,645 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Shannon entropy | 0.23303 |
| G+C content | 0.58659 |
| Mean single sequence MFE | -49.53 |
| Consensus MFE | -34.57 |
| Energy contribution | -35.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 18506525 120 + 22422827 GUUGGACAUUUCCGCAUUCGGUAUGGGGCUGACCAAGUCUGUGGAGCAUAGCUCCUCGCCGGGCACUGCCGGUUCCGUCAUGCUGGAAACUGGGCUGAGAUCCGGGCAAUCCUGGAUAAA .(..(.(((((((((((.(((....(((((.....)))))(.(((((...))))).)(((((......))))).)))..)))).))))).))..)..).(((((((....)))))))... ( -43.20, z-score = -0.08, R) >droYak2.chrX 17134596 115 + 21770863 GCUGGAGAUGUCCGCAUUCGGCAUGGAGCUGACCCAGUCUGAGGAGCACAGCUCCUCGCUGGGCACCACCGGGUCCGACAUGCCGGACGCUGAGCUG-----CGGGCAAUCCUGGAUAAA .(..(.((((((((((((((((.(((.(.((.((((((..(((((((...))))))))))))))).).))).(((((......))))))))))).))-----))))).))))..)..... ( -60.50, z-score = -4.14, R) >droSec1.super_8 810710 120 + 3762037 GUUGGAAUUUUCACCAUUCGGUAUGGGGCUGACCAAGUCUGAGGAGCAUAGCUCCUCGCCGGGCACUGCCGGUUCCGUCAUGCUGGAAUCUGGGAUGAGAUCCGGGCAACCCUGAAAAAA ...(((..(((((((.((((((((((((((.....)))))(((((((...)))))))(((((......))))).....))))))))).....)).))))))))((....))......... ( -44.90, z-score = -1.19, R) >consensus GUUGGAAAUUUCCGCAUUCGGUAUGGGGCUGACCAAGUCUGAGGAGCAUAGCUCCUCGCCGGGCACUGCCGGUUCCGUCAUGCUGGAAACUGGGCUGAGAUCCGGGCAAUCCUGGAUAAA .(..(...........((((((((((((((.....)))))(((((((...)))))))(((((......))))).....)))))))))..(((((......)))))......)..)..... (-34.57 = -35.13 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:27 2011