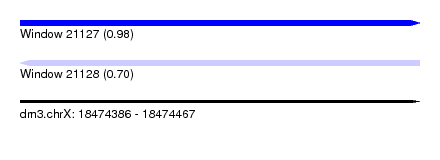
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,474,386 – 18,474,467 |
| Length | 81 |
| Max. P | 0.978484 |

| Location | 18,474,386 – 18,474,467 |
|---|---|
| Length | 81 |
| Sequences | 12 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.72469 |
| G+C content | 0.49012 |
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -7.29 |
| Energy contribution | -7.23 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
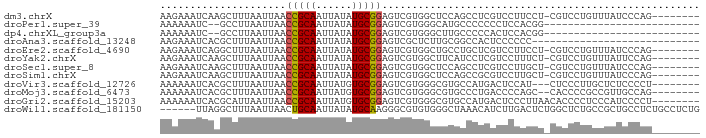
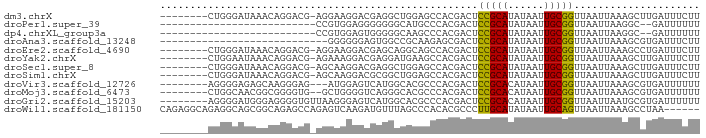
>dm3.chrX 18474386 81 + 22422827 AAGAAAUCAAGCUUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGCUCCAGCCUCGUCCUUCCU-CGUCCUGUUUAUCCCAG-------- ........((((.........(((((......))))).(.((.(((....))).)).)......-......)))).......-------- ( -16.00, z-score = -1.85, R) >droPer1.super_39 425623 62 - 745454 AAAAAAUC--GCCUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGGCAUGCCCCCCCUCCACGG-------------------------- ........--...........(((((......)))((((..(.(((....))))..))))..))-------------------------- ( -16.00, z-score = -1.29, R) >dp4.chrXL_group3a 410140 62 + 2690836 AAAAAAUC--GCCUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGGCUUGCCCCCACUCCACGG-------------------------- ........--...........(((((......)))(((((.(.(((....)))).)))))..))-------------------------- ( -17.40, z-score = -1.99, R) >droAna3.scaffold_13248 1631283 62 + 4840945 AAGAAAUCACGCUUUAAUUAACCGCAAUUAUAUGCGGAGUCGCUCUUGCGGCCACUCCCCCC---------------------------- .....................(((((......))))).(((((....)))))..........---------------------------- ( -14.90, z-score = -2.08, R) >droEre2.scaffold_4690 8787329 81 + 18748788 AAGAAAUCAGGCUUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGCUGCCUGCUCGUCCUUCCU-CGUCCUGUUUAUCCCAG-------- ..((...(((((.........(((((......)))))(((....))))))))....))......-.................-------- ( -17.60, z-score = -1.09, R) >droYak2.chrX 17102106 81 + 21770863 AAGAAAUCAAGCUUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGCUUCAUCCUCGUCCUUUCU-CGUCCUGUUUAUUCCAG-------- .(((((...............(((((......))))).(.((.((......)).)).).)))))-.................-------- ( -12.30, z-score = -0.70, R) >droSec1.super_8 778604 81 + 3762037 AAGAAAUCAAGCUUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGCUCCAGCCUCGUCCUUGCU-CGUCCUGUUUAUCCCAG-------- ..((...((((..........(((((......))))).(.((.(((....))).)).)))))..-..)).............-------- ( -17.20, z-score = -1.59, R) >droSim1.chrX 14263763 81 + 17042790 AAGAAAUCAAGCUUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGCUCCAGCCGCGUCCUUGCU-CGUCCUGUUUAUCCCAG-------- ..((...((((..........(((((......))))).(.((((((....)))))).)))))..-..)).............-------- ( -20.80, z-score = -2.37, R) >droVir3.scaffold_12726 2303719 79 + 2840439 AAAAAAUCACGCUUUAAUUAACCGCAAUUAUGUGCGGAGUCGUGGGCGUGCCAUGACUCCAU---CUCCCUUGCUCUCCCCU-------- ..........((...........(((......)))((((((((((.....))))))))))..---.......))........-------- ( -22.30, z-score = -3.33, R) >droMoj3.scaffold_6473 10753063 80 - 16943266 AAAAAAUCACGCUUUAAUUAACCGCAAUUAUGUGCGGAGUCGUGGGCGUGCCCUGACCCCAGC--CACCCCGCCGUUGCCAG-------- ..........((.........(((((......)))))....(((((.(((..(((....))).--))))))))....))...-------- ( -21.80, z-score = -1.16, R) >droGri2.scaffold_15203 1255669 82 + 11997470 AAAAAAUCACGCAUUAAUUAACCGCAAUUAUGUGCGGAGUCGUGGGCGUGCCAUGACUCCCUUAACACCCCUCCCAUCCCCU-------- ..........(((((((((......))))).))))((((((((((.....))))))))))......................-------- ( -22.30, z-score = -3.41, R) >droWil1.scaffold_181150 1471278 84 + 4952429 ------UUAGGCUUUAAUUAACUGCAAUUAUAUGCAAGGGCGUGUGGGCUAAACAUCUUGACUCUGGCUCUGCCGCUGCCUCUGCCUCUG ------..((((..........((((......))))((((((((.((((((.............))))))...))).))).))))))... ( -21.22, z-score = -0.29, R) >consensus AAGAAAUCAAGCUUUAAUUAACCGCAAUUAUAUGCGGAGUCGUGGGUGCGCCCUCGCCCCACCU_CGUCCUGUUUAUCCCAG________ .....................(((((......)))))..................................................... ( -7.29 = -7.23 + -0.07)
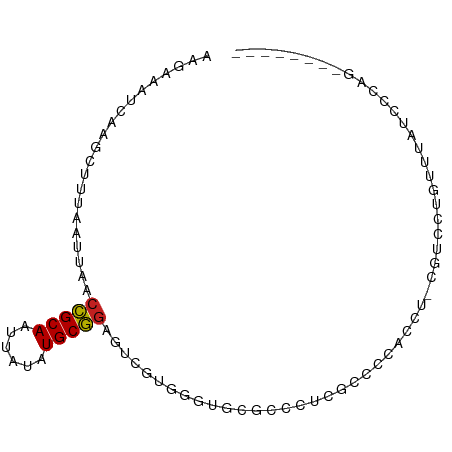
| Location | 18,474,386 – 18,474,467 |
|---|---|
| Length | 81 |
| Sequences | 12 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.72469 |
| G+C content | 0.49012 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -7.31 |
| Energy contribution | -7.08 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
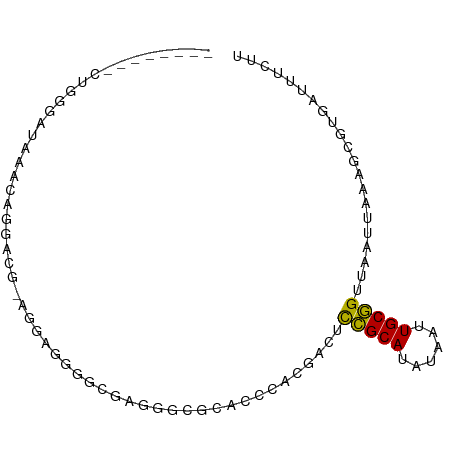
>dm3.chrX 18474386 81 - 22422827 --------CUGGGAUAAACAGGACG-AGGAAGGACGAGGCUGGAGCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAAGCUUGAUUUCUU --------(((.......)))..((-((......((.(((....))).))...(((((......)))))..........))))....... ( -19.30, z-score = -1.06, R) >droPer1.super_39 425623 62 + 745454 --------------------------CCGUGGAGGGGGGGCAUGCCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAGGC--GAUUUUUU --------------------------..((((((..((((....))).)..))))))....((((((...........))--)))).... ( -20.40, z-score = -1.46, R) >dp4.chrXL_group3a 410140 62 - 2690836 --------------------------CCGUGGAGUGGGGGCAAGCCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAGGC--GAUUUUUU --------------------------..(((((((.((((....))).).)))))))....((((((...........))--)))).... ( -22.70, z-score = -2.60, R) >droAna3.scaffold_13248 1631283 62 - 4840945 ----------------------------GGGGGGAGUGGCCGCAAGAGCGACUCCGCAUAUAAUUGCGGUUAAUUAAAGCGUGAUUUCUU ----------------------------..(.(((((.(((....).)).))))).)....(((..((.((.....)).))..))).... ( -21.80, z-score = -2.47, R) >droEre2.scaffold_4690 8787329 81 - 18748788 --------CUGGGAUAAACAGGACG-AGGAAGGACGAGCAGGCAGCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAAGCCUGAUUUCUU --------(((.......)))....-((((((......(((((..........(((((......))))).........))))).)))))) ( -20.11, z-score = -1.14, R) >droYak2.chrX 17102106 81 - 21770863 --------CUGGAAUAAACAGGACG-AGAAAGGACGAGGAUGAAGCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAAGCUUGAUUUCUU --------(((.......)))...(-(((((...((.((......)).))...(((((......)))))...............)))))) ( -15.80, z-score = -0.89, R) >droSec1.super_8 778604 81 - 3762037 --------CUGGGAUAAACAGGACG-AGCAAGGACGAGGCUGGAGCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAAGCUUGAUUUCUU --------(((.......)))..((-(((.....((.(((....))).))...(((((......))))).........)))))....... ( -23.10, z-score = -2.32, R) >droSim1.chrX 14263763 81 - 17042790 --------CUGGGAUAAACAGGACG-AGCAAGGACGCGGCUGGAGCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAAGCUUGAUUUCUU --------(((.......)))..((-(((.....((.(((....))).))...(((((......))))).........)))))....... ( -23.90, z-score = -2.20, R) >droVir3.scaffold_12726 2303719 79 - 2840439 --------AGGGGAGAGCAAGGGAG---AUGGAGUCAUGGCACGCCCACGACUCCGCACAUAAUUGCGGUUAAUUAAAGCGUGAUUUUUU --------................(---.(((((((.(((.....))).))))))))....(((..((.((.....)).))..))).... ( -18.70, z-score = -0.20, R) >droMoj3.scaffold_6473 10753063 80 + 16943266 --------CUGGCAACGGCGGGGUG--GCUGGGGUCAGGGCACGCCCACGACUCCGCACAUAAUUGCGGUUAAUUAAAGCGUGAUUUUUU --------(((((..((((......--))))..)))))..(((((........(((((......))))).........)))))....... ( -28.73, z-score = -1.61, R) >droGri2.scaffold_15203 1255669 82 - 11997470 --------AGGGGAUGGGAGGGGUGUUAAGGGAGUCAUGGCACGCCCACGACUCCGCACAUAAUUGCGGUUAAUUAAUGCGUGAUUUUUU --------.((((.(.(..((.((((((.(.....).)))))).))..).)))))(((..((((((....)))))).))).......... ( -20.40, z-score = 0.15, R) >droWil1.scaffold_181150 1471278 84 - 4952429 CAGAGGCAGAGGCAGCGGCAGAGCCAGAGUCAAGAUGUUUAGCCCACACGCCCUUGCAUAUAAUUGCAGUUAAUUAAAGCCUAA------ ...((((((.(((...(((.((((............)))).))).....)))))((((......))))..........))))..------ ( -19.50, z-score = -0.45, R) >consensus ________CUGGGAUAAACAGGACG_AGGAGGGGCGAGGGCGCACCCACGACUCCGCAUAUAAUUGCGGUUAAUUAAAGCGUGAUUUCUU .....................................................(((((......)))))..................... ( -7.31 = -7.08 + -0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:22 2011