
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,471,091 – 18,471,189 |
| Length | 98 |
| Max. P | 0.981207 |

| Location | 18,471,091 – 18,471,189 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.03 |
| Shannon entropy | 0.37290 |
| G+C content | 0.43826 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -16.71 |
| Energy contribution | -16.66 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18471091 98 - 22422827 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUG-CGCCCGCACAAAGCAAAAGGGGCAGCUCGAGCCCCAG---------- (((((((((....))))..(((......((......))((((((((((....)))))))))-)))).)))))........(((((.......)))))..---------- ( -33.90, z-score = -3.67, R) >droSim1.chrX 14260506 98 - 17042790 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUG-CGCCCGCACAAAGCAAAAGGGGCAGCUCGAGCCCCAG---------- (((((((((....))))..(((......((......))((((((((((....)))))))))-)))).)))))........(((((.......)))))..---------- ( -33.90, z-score = -3.67, R) >droSec1.super_8 775313 98 - 3762037 UGUGUUUGUGUAAGCAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUG-CGCCCGCACAAAGCAAAAGGGGCAGCCCGAGCCGCAG---------- (((((((((((..((.....((....))..........((((((((((....)))))))))-)))..)))))))((....(((....)))..)))))).---------- ( -31.00, z-score = -2.10, R) >droYak2.chrX 17098736 108 - 21770863 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUG-CGCCCGCACAAAGCAAAAGGGGCAGCACGAGCAGCAGGACGAAGGAU (((((((((((.........(((..(((((......)))))(((((((....)))))))))-)((((((.....)).....)))).))))))))).))...(....).. ( -32.30, z-score = -2.71, R) >droEre2.scaffold_4690 8784373 85 - 18748788 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGCAGGAGAUUAAAAAGUUUCCUG-CGCCCGCAC-AAGCAAAAGGGGCAG---------------------- ..(((((((((........(((......((......))((((((((((....)))))))))-)))).))))-)))))..........---------------------- ( -27.70, z-score = -3.12, R) >droAna3.scaffold_13248 1615304 79 - 4840945 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUG-CGCCCGCACAAAAGCAGG----------------------------- (((.(((((((........(((...(((((......))))).((((((....))))))...-.))).))))))).)))..----------------------------- ( -19.40, z-score = -1.82, R) >droWil1.scaffold_181150 1463979 82 - 4952429 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAGGCCGCAUAUACAUAGAUAUCUG--------------------------- .(((((((((((......((((...(((((......))))).((((((....))))))..))))....)))))))))))...--------------------------- ( -21.30, z-score = -3.20, R) >droMoj3.scaffold_6473 10746891 85 + 16943266 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUG-CGCCCGCACUGUACGACGGGAGCG----------------------- ...((((((....))))))(((...(((((......))))).((((((....))))))...-.)))(((.(((.....)))..)))----------------------- ( -21.20, z-score = -0.81, R) >droGri2.scaffold_15203 1251702 85 - 11997470 UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUG-CGCCCGCACAGCACAAUAAGAUCG----------------------- ((((((.((((........(((...(((((......))))).((((((....))))))...-.))).)))))))))).........----------------------- ( -20.40, z-score = -1.74, R) >consensus UGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUG_CGCCCGCACAAAGCAAAAGGGGCAG______________________ (((((((((....))))..(((...(((((......))))).((((((....)))))).....))).)))))..................................... (-16.71 = -16.66 + -0.06)

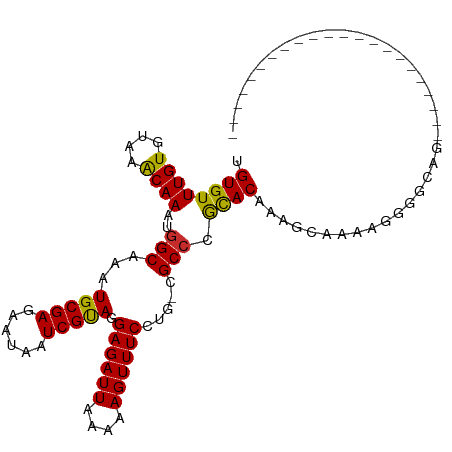
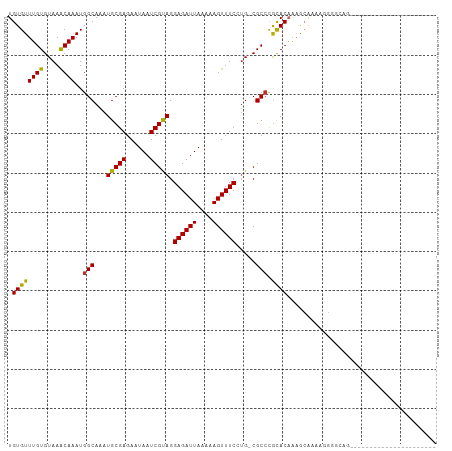
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:20 2011