
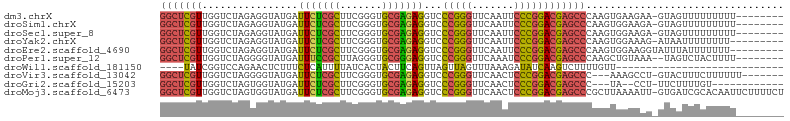
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,459,796 – 18,459,891 |
| Length | 95 |
| Max. P | 0.998562 |

| Location | 18,459,796 – 18,459,891 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.60942 |
| G+C content | 0.51058 |
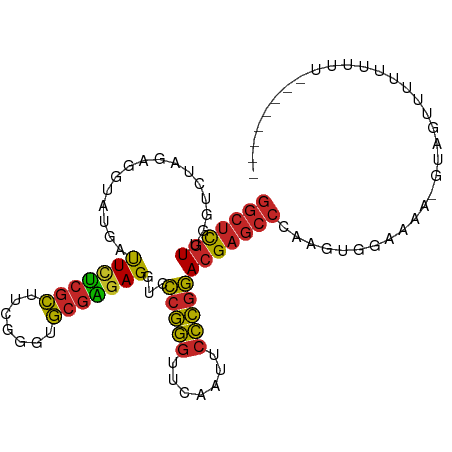
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.07 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18459796 95 + 22422827 GGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUGAAGAA-GUAGUUUUUUUUU-------- (((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).....((((((-(....))))))).-------- ( -32.40, z-score = -1.33, R) >droSim1.chrX 14249098 95 + 17042790 GGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUGGAAGA-GUAGUUUUUUUUU-------- (((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).....((((((-(....))))))).-------- ( -32.00, z-score = -0.91, R) >droSec1.super_8 764366 95 + 3762037 GGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUGGAAGA-GUAGUUUUUUUUU-------- (((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).....((((((-(....))))))).-------- ( -32.00, z-score = -0.91, R) >droYak2.chrX 17087742 94 + 21770863 GGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUGGAAAG-AUAAUUUUUUUU--------- (((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).....((((((-(....)))))))--------- ( -32.20, z-score = -1.14, R) >droEre2.scaffold_4690 8773556 95 + 18748788 GGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUGGAAGGUAUUUAUUUUUUU--------- (((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).(((((((.....)))))))....--------- ( -31.80, z-score = -0.93, R) >droPer1.super_12 1247070 94 - 2414086 GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGCUGUAAA--UAGUCUACUUUU-------- ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))...((((....--))))........-------- ( -32.60, z-score = -0.60, R) >droWil1.scaffold_181150 1448896 72 + 4952429 ----UAUCGGUCCAGAACUCUUUCUCAUUUUAUCACUACUUCAGUUAGUUAGUUUAAAGAUAUCAAGUCUUUUGUU---------------------------- ----........(((((..(((.....(((((..((((((......)).)))).))))).....)))..)))))..---------------------------- ( -6.10, z-score = 0.48, R) >droVir3.scaffold_13042 3405262 93 + 5191987 GGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCC---AAAGCCU-GUACUUUCUUUUUU------- ((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).---.......-..............------- ( -33.20, z-score = -0.78, R) >droGri2.scaffold_15203 1232686 86 + 11997470 GGCUCGUUGGUCUAGUGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCC---UA--CCU-UUCUUUUGU------------ (((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).---..--...-.........------------ ( -31.50, z-score = -1.17, R) >droMoj3.scaffold_6473 10728418 103 - 16943266 GGCUCGUUGGUCUAGUGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCCGCUUAAAAUU-GUGAUCGCACAAUUCUUUUCU (((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).......((((-(((....)))))))....... ( -36.40, z-score = -1.68, R) >consensus GGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUGGAAAA_GUAGUUUUUUUUU________ (((((((................(((((((.......)))))))...(((((.......))))))))))))................................. (-27.39 = -27.07 + -0.32)
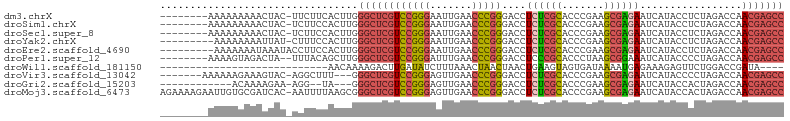
| Location | 18,459,796 – 18,459,891 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Shannon entropy | 0.60942 |
| G+C content | 0.51058 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.998562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18459796 95 - 22422827 --------AAAAAAAAACUAC-UUCUUCACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCC --------.............-...........((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -29.20, z-score = -3.03, R) >droSim1.chrX 14249098 95 - 17042790 --------AAAAAAAAACUAC-UCUUCCACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCC --------.............-...........((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -29.20, z-score = -2.99, R) >droSec1.super_8 764366 95 - 3762037 --------AAAAAAAAACUAC-UCUUCCACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCC --------.............-...........((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -29.20, z-score = -2.99, R) >droYak2.chrX 17087742 94 - 21770863 ---------AAAAAAAAUUAU-CUUUCCACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCC ---------............-...........((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -29.20, z-score = -2.77, R) >droEre2.scaffold_4690 8773556 95 - 18748788 ---------AAAAAAAUAAAUACCUUCCACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCC ---------........................((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -29.20, z-score = -3.13, R) >droPer1.super_12 1247070 94 + 2414086 --------AAAAGUAGACUA--UUUACAGCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC --------....(((((...--)))))......((((((((((((.......)))))......((((.......))))...................))))))) ( -27.20, z-score = -1.40, R) >droWil1.scaffold_181150 1448896 72 - 4952429 ----------------------------AACAAAAGACUUGAUAUCUUUAAACUAACUAACUGAAGUAGUGAUAAAAUGAGAAAGAGUUCUGGACCGAUA---- ----------------------------.......(((((....((((.......((((.(....)))))........))))..)))))...........---- ( -6.36, z-score = 0.79, R) >droVir3.scaffold_13042 3405262 93 - 5191987 -------AAAAAAGAAAGUAC-AGGCUUU---GGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCC -------.......(((((..-..)))))---.((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -31.20, z-score = -2.01, R) >droGri2.scaffold_15203 1232686 86 - 11997470 ------------ACAAAAGAA-AGG--UA---GGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCACUAGACCAACGAGCC ------------.........-...--..---.((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -29.20, z-score = -2.14, R) >droMoj3.scaffold_6473 10728418 103 + 16943266 AGAAAAGAAUUGUGCGAUCAC-AAUUUUAAGCGGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCACUAGACCAACGAGCC .....(((((((((....)))-)))))).....((((((((((((.......)))))((..((((((.......)))))).))..............))))))) ( -35.80, z-score = -3.14, R) >consensus ________AAAAAAAAACUAC_UCUUCCACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCC .................................((((((((((((.......)))))....((((((.......)))))).................))))))) (-24.04 = -24.18 + 0.14)
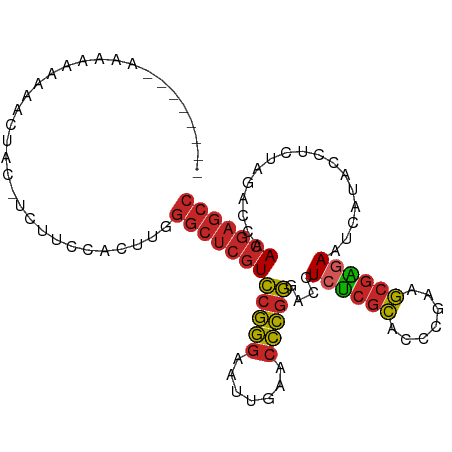
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:19 2011