| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,443,937 – 18,444,046 |
| Length | 109 |
| Max. P | 0.991426 |

| Location | 18,443,937 – 18,444,046 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Shannon entropy | 0.40628 |
| G+C content | 0.37751 |
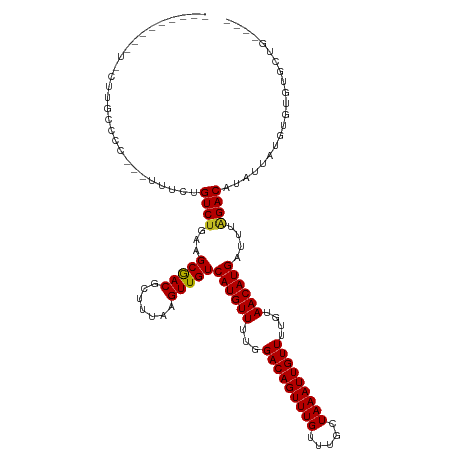
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
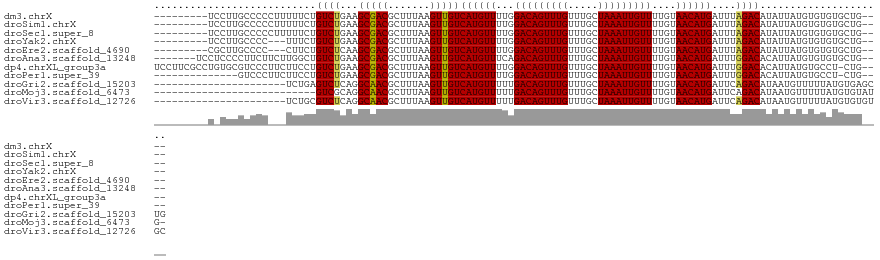
>dm3.chrX 18443937 109 - 22422827 ---------UCCUUGCCCCCUUUUUCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUAGACAUAUUAUGUGUGUGCUG---- ---------.....((((.(......(((((((((((((.......)))))((((((..((((((((((.....))))))))))...)))))).))))))))......).).).))..---- ( -24.90, z-score = -2.43, R) >droSim1.chrX 14239804 109 - 17042790 ---------UCCUUGCCCCCUUUUUCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUAGACAUAUUAUGUGUGUGCUG---- ---------.....((((.(......(((((((((((((.......)))))((((((..((((((((((.....))))))))))...)))))).))))))))......).).).))..---- ( -24.90, z-score = -2.43, R) >droSec1.super_8 749966 109 - 3762037 ---------UCCUUGCCCCCUUUUUCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUAGACAUAUUAUGUGUGUGCUG---- ---------.....((((.(......(((((((((((((.......)))))((((((..((((((((((.....))))))))))...)))))).))))))))......).).).))..---- ( -24.90, z-score = -2.43, R) >droYak2.chrX 17072944 106 - 21770863 ---------UCCUUGCCCC---UUUCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUAGACAUAUUAUGUGUGUGCUG---- ---------..........---....(((((((((((((.......)))))((((((..((((((((((.....))))))))))...)))))).))))))))................---- ( -24.70, z-score = -2.12, R) >droEre2.scaffold_4690 8759683 106 - 18748788 ---------CGCUUGCCCC---CUUCUGUCUCAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUAGACAUAUUAUGUGUGUGCUG---- ---------((((((....---.........))))))((((..((.(((((((((((..((((((((((.....))))))))))...))))))))...))).))....))))......---- ( -25.12, z-score = -1.73, R) >droAna3.scaffold_13248 1589675 111 - 4840945 -------UCCUCCCCUUCUUCUUGGCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUCAGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUGGACACAUUAUGUGUGUGCUG---- -------................((((((((((((((((((.....).))))..)))))))))))))......((((((..(........)..))))))((((((...))))))....---- ( -28.90, z-score = -2.37, R) >dp4.chrXL_group3a 374675 117 - 2690836 UCCUUCGCCUGUGCGUCCCUUCUUCCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUGGACACAUUAUGUGCCU-CUG---- ............(((((.((((.........)))).)))))........((((((((..((((((((((.....))))))))))...))))))))..((.(((.....))))).-...---- ( -32.00, z-score = -3.48, R) >droPer1.super_39 390046 103 + 745454 --------------GUCCCUUCUUCCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUGGACACAUUAUGUGCCU-CUG---- --------------(((.((((.........)))).)))..........((((((((..((((((((((.....))))))))))...))))))))..((.(((.....))))).-...---- ( -26.70, z-score = -2.73, R) >droGri2.scaffold_15203 1216324 100 - 11997470 ----------------------UCUGAGUCUCAGGCAACGCUUUAAGUUGUCAUGUUUUUGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUCAGACAUAAUGUUUUUAUGUGAGCUG ----------------------.(((((((...((((((.......))))))(((((...(((((((((.....)))))))))....))))))))))))((((((.....))))))...... ( -29.10, z-score = -3.00, R) >droMoj3.scaffold_6473 10713353 94 + 16943266 ---------------------------GUCGCAGGCAACGCUUUAAGUUGUCAUGUUUUUGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUCAGACAUAAUGUUUUUAUGUGUAUG- ---------------------------...((.(....))).....(..((((((((...(((((((((.....)))))))))....)))))))).)(.((((((.....)))))).)...- ( -21.30, z-score = -1.02, R) >droVir3.scaffold_12726 2266517 100 - 2840439 ----------------------UCUGCGUCUCAGGCAACGCUUUAAGUUGUCAUGUUUUUGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUCAGACAUAAUGUUUUUAUGUGUGUGC ----------------------.(((.(((...((((((.......))))))(((((...(((((((((.....)))))))))....)))))))).)))((((((.....))))))...... ( -25.00, z-score = -1.55, R) >consensus _________U_CUUGCCCC___UUUCUGUCUGAAGCGACGCUUUAAGUUGUCAUGUUUUGGACAGUUUGUUUGCUAAAUUGUUUUGUAACAUGAUUUAGACAUAUUAUGUGUGUGCUG____ ...........................((((...(((((.......)))))((((((...(((((((((.....)))))))))....))))))....))))..................... (-19.31 = -19.03 + -0.28)
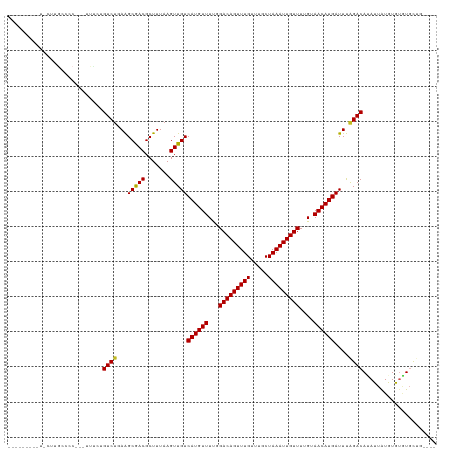
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:17 2011