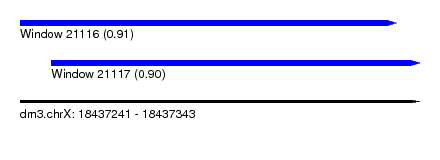
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,437,241 – 18,437,343 |
| Length | 102 |
| Max. P | 0.910328 |

| Location | 18,437,241 – 18,437,337 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 68.25 |
| Shannon entropy | 0.43687 |
| G+C content | 0.30314 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -10.25 |
| Energy contribution | -9.70 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
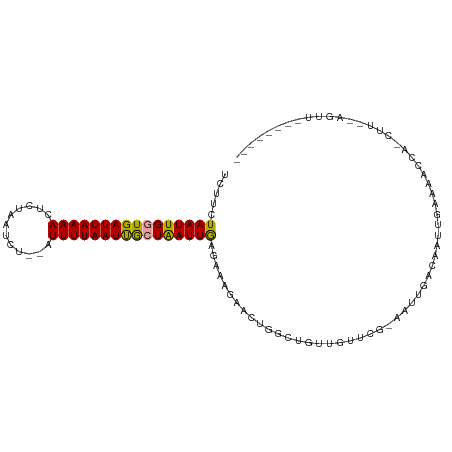
>dm3.chrX 18437241 96 + 22422827 ACACACAUUCUUCUAAUUGGUGAUUAAAACUCUAAUCCUGUUUUUAAUUGCUAAUUAAGAAAGAACUGGCUGUUGUUCGCA-UUGACAAUUGAAAGC---------- .......(((((((((((((..((((((((.........).)))))))..)))))).)).)))))((..(.((((((....-..)))))).)..)).---------- ( -19.40, z-score = -1.82, R) >droYak2.chrX 17066132 97 + 21770863 ACACACAUUCAGCUAAUUGCUGAUUAAAACUCAAUUCUA--UUUUAAUCAAUGAUUGAGAGGGAACUGAUUGCUCU----AAUUGACAAUUCAAAAUCAGCAU---- ........(((((.....))))).......(((.((((.--((((((((...)))))))).)))).))).((((..----..((((....))))....)))).---- ( -20.50, z-score = -2.39, R) >droSec1.super_8 743336 101 + 3762037 ----ACACACUUCUAAUUGGUGAUUAAAAAUAUAAUCUA--UUUUAAUUGCUAAUUGAGAAAGAACUGGCUGUUGUUCGAAAUUAACAAUCGAAAUCCAUCUUAGUU ----......((((((((((..(((((((..........--)))))))..)))))).))))..((((((.((...(((((.........)))))...)).).))))) ( -18.80, z-score = -2.11, R) >consensus ACACACAUUCUUCUAAUUGGUGAUUAAAACUCUAAUCUA__UUUUAAUUGCUAAUUGAGAAAGAACUGGCUGUUGUUCG_AAUUGACAAUUGAAAACCA_C_U____ .............((((((((((((((((............)))))))))))))))).................................................. (-10.25 = -9.70 + -0.55)


| Location | 18,437,249 – 18,437,343 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.57 |
| Shannon entropy | 0.42274 |
| G+C content | 0.29784 |
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -10.38 |
| Energy contribution | -9.83 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18437249 94 + 22422827 UCUUCUAAUUGGUGAUUAAAACUCUAAUCCUGUUUUUAAUUGCUAAUUAAGAAAGAACUGGCUGUUGUUCGCA-UUGACAAUUGAAAGC----UU--AGUU-------- ..((((((((((..((((((((.........).)))))))..)))))).))))..((((((((((((((....-..))))))....)))----.)--))))-------- ( -21.00, z-score = -2.45, R) >droYak2.chrX 17066140 95 + 21770863 UCAGCUAAUUGCUGAUUAAAACUCAAUUCU--AUUUUAAUCAAUGAUUGAGAGGGAACUGAUUGCUCU----AAUUGACAAUUCAAAAUCAGCAUGCAGUU-------- ...((....((((((((.....(((.((((--.((((((((...)))))))).)))).)))(((.((.----....))))).....)))))))).))....-------- ( -22.90, z-score = -2.20, R) >droSec1.super_8 743341 104 + 3762037 -CUUCUAAUUGGUGAUUAAAAAUAUAAUCU--AUUUUAAUUGCUAAUUGAGAAAGAACUGGCUGUUGUUCGAAAUUAACAAUCGAAAUCCAUCUU--AGUUAACUAGUU -.((((((((((..(((((((.........--.)))))))..)))))).))))..((((((.((...(((((.........)))))...)).).)--))))........ ( -19.50, z-score = -2.15, R) >consensus UCUUCUAAUUGGUGAUUAAAACUCUAAUCU__AUUUUAAUUGCUAAUUGAGAAAGAACUGGCUGUUGUUCG_AAUUGACAAUUGAAAACCA_CUU__AGUU________ .....((((((((((((((((............))))))))))))))))............................................................ (-10.38 = -9.83 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:13 2011