| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,433,146 – 18,433,227 |
| Length | 81 |
| Max. P | 0.940690 |

| Location | 18,433,146 – 18,433,227 |
|---|---|
| Length | 81 |
| Sequences | 11 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Shannon entropy | 0.18741 |
| G+C content | 0.31530 |
| Mean single sequence MFE | -13.65 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.940690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18433146 81 - 22422827 --UCCCUUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAU-GAAAACGACCACAAAA --..........((((((((.(((......))).)))))))).......(((((............)-))))............ ( -12.60, z-score = -2.25, R) >droSim1.chrX 14228827 81 - 17042790 --UCCCUUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAU-GAAAACGACCACAAAA --..........((((((((.(((......))).)))))))).......(((((............)-))))............ ( -12.60, z-score = -2.25, R) >droSec1.super_8 739307 81 - 3762037 --UCCCUUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAU-GAAAACGACCACAAAA --..........((((((((.(((......))).)))))))).......(((((............)-))))............ ( -12.60, z-score = -2.25, R) >droYak2.chrX 17061958 81 - 21770863 --UCCCUUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAU-GAAAACGACCACAAAA --..........((((((((.(((......))).)))))))).......(((((............)-))))............ ( -12.60, z-score = -2.25, R) >droEre2.scaffold_4690 8748799 81 - 18748788 --UCCCUUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAU-GAAAACGACCACAAAA --..........((((((((.(((......))).)))))))).......(((((............)-))))............ ( -12.60, z-score = -2.25, R) >droAna3.scaffold_13248 1576904 81 - 4840945 --UCCCUUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAU-GAAAACGACCACAAAA --..........((((((((.(((......))).)))))))).......(((((............)-))))............ ( -12.60, z-score = -2.25, R) >dp4.chrXL_group3a 363534 82 - 2690836 --UCCACUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAUGGAAAACGACCACAAAA --((((......((((((((.(((......))).))))))))....(((((.......)))))...)))).............. ( -14.60, z-score = -2.28, R) >droPer1.super_39 378192 82 + 745454 --UCCACUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCUAACCAAUUAUGAUGGAAAACGACCACAUAA --..........((((((((.(((......))).))))))))..................(((((.(((.......)))))))) ( -16.30, z-score = -2.54, R) >droWil1.scaffold_181150 1421077 82 - 4952429 GCC--GGAGUAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAGACCAAUUAUAAAAGAAAACGAACACAAAA ..(--(......((((((((.(((......))).)))))))).......((((...............)))).))......... ( -12.46, z-score = -1.23, R) >droVir3.scaffold_12726 2250038 83 - 2840439 GCCCGGGGGUAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAU-AAAGAAAACGACCACGAAA ...((..(((..((((((((.(((......))).)))))))).......((((...........-...))))...))).))... ( -16.44, z-score = -0.94, R) >droGri2.scaffold_15203 1198979 82 - 11997470 GCCC-GGGGUAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAU-AAAGAAAACGACCACAAAC ....-..(((..((((((((.(((......))).)))))))).......((((...........-...))))...)))...... ( -14.74, z-score = -1.05, R) >consensus __UCCCUUAAAAGCCAAUUAAAUGUGAGAUCGUGUAAUUGGCAAAUUAAUUUCAAACCAAUUAUAAU_GAAAACGACCACAAAA ............((((((((.(((......))).)))))))).......................................... (-12.10 = -12.10 + -0.00)

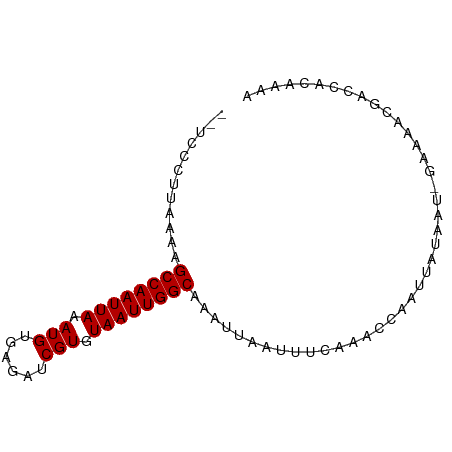
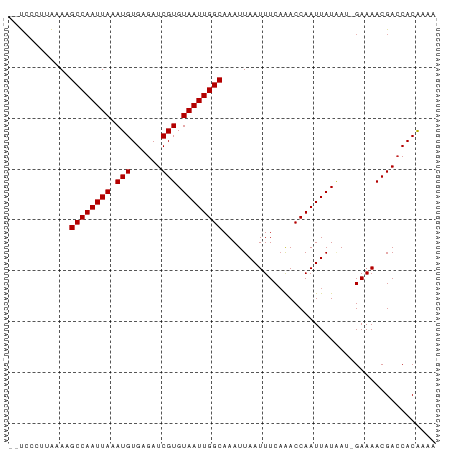
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:11 2011