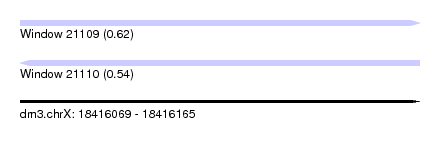
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,416,069 – 18,416,165 |
| Length | 96 |
| Max. P | 0.618201 |

| Location | 18,416,069 – 18,416,165 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 61.14 |
| Shannon entropy | 0.62418 |
| G+C content | 0.51679 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
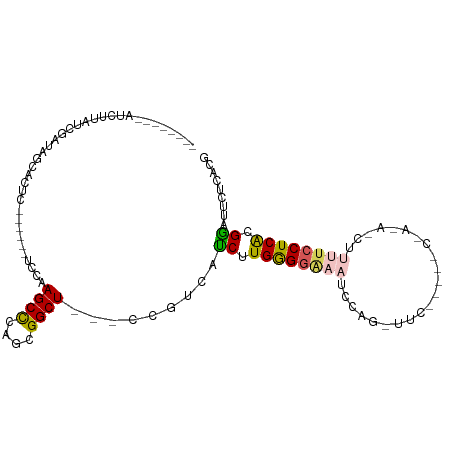
>dm3.chrX 18416069 96 + 22422827 --------AUAUUUUCGAUAGCACUC------UUCAAGCCCAACGGCUUCGCCGUCAUCUUGAGGCAAUCCAGCUUCGGAUACAAGAACUUUUCCUCACGGAUUCUCACG --------.....(((((.(((...(------((((((....(((((...)))))...))))))).......))))))))....((((....(((....))))))).... ( -20.60, z-score = -1.59, R) >droSim1.chrX_random 4768511 72 + 5698898 --------AUUUCAUCGACAGCACUC------UCCAAGCCCAGCGGCU---CCGUUAUCUUGGGGAAACUC---------------------UCCUCACGGAUUCUCACG --------................((------((((((...((((...---.))))..)))))))).....---------------------(((....)))........ ( -16.30, z-score = -1.28, R) >droSec1.super_8 722233 72 + 3762037 --------AUUUUAUCGACAGCACUC------UCCAAGCCCAGCGGCU---CCGUUAUCUUGGGGAAAUUC---------------------UCCUCACGGAUUCUCACG --------................((------((((((...((((...---.))))..)))))))).....---------------------(((....)))........ ( -16.30, z-score = -1.34, R) >droYak2.chrX 17046264 107 + 21770863 UAUCUGGGGUCACCUCGCUUGCAUUCACCUUCUCCAAGCUCUUUGGCU---CCGCCGUCUUGGGCAAAUCCAGUUUCAGAACCGACAACUUUUCCUCACGAAUUCUUACC ..((((((((((....(((((.............)))))....)))))---))(((......)))............))).............................. ( -20.02, z-score = 0.09, R) >droEre2.scaffold_4690 8732540 101 + 18748788 CAUCUGCGGACUUCUCGUUUACAUUA------UCCAAGCUCUUCGGCU---CCGCCACCUUGGGGGAAUCCAGCUUCGAGACCGAAAGCUUUUCCUCGCGGAUUCUCACG .((((((((.................------((((((.....((...---.))....))))))((((...(((((((....)).))))).))))))))))))....... ( -27.10, z-score = -0.78, R) >consensus ________AUCUUAUCGAUAGCACUC______UCCAAGCCCAGCGGCU___CCGUCAUCUUGGGGAAAUCCAG_UUC_____C_A_A_CUUUUCCUCACGGAUUCUCACG ................................((((((....((((.....))))...))))))............................(((....)))........ ( -9.24 = -9.28 + 0.04)

| Location | 18,416,069 – 18,416,165 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 61.14 |
| Shannon entropy | 0.62418 |
| G+C content | 0.51679 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
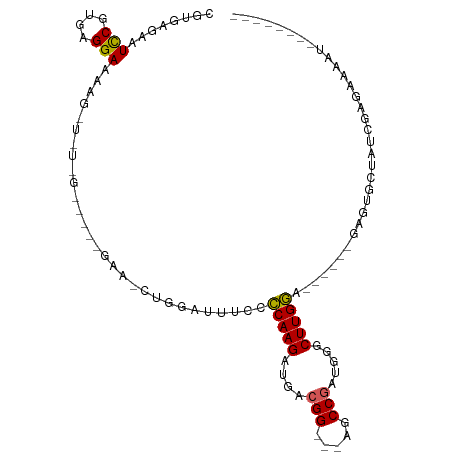
>dm3.chrX 18416069 96 - 22422827 CGUGAGAAUCCGUGAGGAAAAGUUCUUGUAUCCGAAGCUGGAUUGCCUCAAGAUGACGGCGAAGCCGUUGGGCUUGAA------GAGUGCUAUCGAAAAUAU-------- .(..(((((((....)))....))))..)...((((((.((....))(((((.(((((((...)))))))..))))).------....))).))).......-------- ( -28.00, z-score = -1.68, R) >droSim1.chrX_random 4768511 72 - 5698898 CGUGAGAAUCCGUGAGGA---------------------GAGUUUCCCCAAGAUAACGG---AGCCGCUGGGCUUGGA------GAGUGCUGUCGAUGAAAU-------- (....)..(((....)))---------------------.((((((.(((((.((.((.---...)).))..))))).------))).)))...........-------- ( -18.00, z-score = -0.18, R) >droSec1.super_8 722233 72 - 3762037 CGUGAGAAUCCGUGAGGA---------------------GAAUUUCCCCAAGAUAACGG---AGCCGCUGGGCUUGGA------GAGUGCUGUCGAUAAAAU-------- (....)..(((....)))---------------------((..(((.(((((.((.((.---...)).))..))))).------))).....))........-------- ( -17.10, z-score = -0.22, R) >droYak2.chrX 17046264 107 - 21770863 GGUAAGAAUUCGUGAGGAAAAGUUGUCGGUUCUGAAACUGGAUUUGCCCAAGACGGCGG---AGCCAAAGAGCUUGGAGAAGGUGAAUGCAAGCGAGGUGACCCCAGAUA ((......(((....)))......((((.(((......(((.((((((......)))))---).)))....(((((.(.........).)))))))).)))).))..... ( -27.60, z-score = -0.24, R) >droEre2.scaffold_4690 8732540 101 - 18748788 CGUGAGAAUCCGCGAGGAAAAGCUUUCGGUCUCGAAGCUGGAUUCCCCCAAGGUGGCGG---AGCCGAAGAGCUUGGA------UAAUGUAAACGAGAAGUCCGCAGAUG .(((.((.(((....))).(((((((((((..((..(((((......))..)))..)).---.)))).)))))))...------................)))))..... ( -31.30, z-score = -0.78, R) >consensus CGUGAGAAUCCGUGAGGAAAAG_U_U_G_____GAA_CUGGAUUUCCCCAAGAUGACGG___AGCCGAUGGGCUUGGA______GAGUGCUAUCGAGAAAAU________ ........(((....))).............................(((((....(((.....))).....)))))................................. (-12.72 = -12.44 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:07 2011