| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,405,110 – 18,405,227 |
| Length | 117 |
| Max. P | 0.843884 |

| Location | 18,405,110 – 18,405,227 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.36 |
| Shannon entropy | 0.31879 |
| G+C content | 0.47060 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -18.71 |
| Energy contribution | -19.03 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.512490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
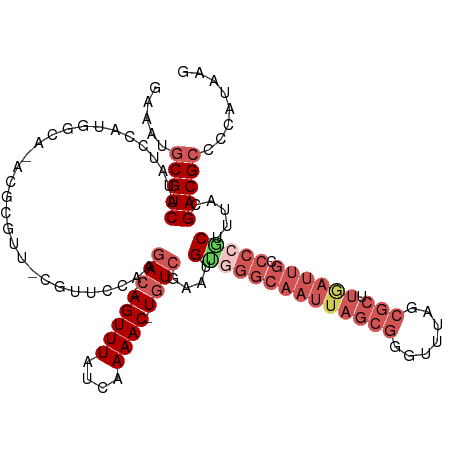
>dm3.chrX 18405110 117 + 22422827 CUUAUGGGGCGUCGUAAAGUGGGGCAAUCAAGCGCUAAACCCGCUAAUUGCCCACAUUCGACA-GUUUUGAUAAACUUGUCUUGGAACA-AACGCAU-UGCCAUGGAUAUGACGCAUUUC ........((((((((.(((((((((((..((((.......)))).))))))).)))).((((-((((....)))).)))).(((..((-(.....)-)))))....))))))))..... ( -34.80, z-score = -1.82, R) >droEre2.scaffold_4690 8722314 94 + 18748788 CUUAUGGGCCGUCGUAAAG------------------------CUAAUUGCCGACAUUCGACAAGUUUUGAUAAACU-CUUUUGCAAACGAACGCGU-UGCCAUGGGUAUGACGCAUUUC .......((.(((((((((------------------------((...((.((.....)).)))))))).....(((-(....((((.((....)))-)))...)))))))))))..... ( -17.90, z-score = 1.07, R) >droYak2.chrX 17036162 118 + 21770863 CUUAUGGGGCGUCGUAAAGCGGGGCAAUUAGGCGCUAAACCCGCUAAUUGCCGGCAUUCGACA-GUUUUGAUAAACU-GUCUUGGAGUAUAACGAAUAUAACGUGGAUAUGACGCAUUUC ........((((((((..(((.(((((((((.((.......)))))))))))...((((((((-((((....)))))-)))..(........)))))....)))...))))))))..... ( -39.60, z-score = -3.03, R) >droSec1.super_8 711306 115 + 3762037 CUUAUGGGGCGUCGUAAAGCGGGGCAAUCAAGCGCUAAACCCGCUAAUUGCCCACAUUCGACA-GUUUUGAUAAACU-GUCUUGGAACG-AACGCGU--UGCCUGGAUAUGACGCAUUUC ........((((((((.....(((((((..((((.......)))).)))))))...(((((((-((((....)))))-)))..((((((-....)))--).)).)))))))))))..... ( -41.20, z-score = -3.09, R) >droSim1.chrX 14210672 116 + 17042790 CUUAUGGGGCGUCGUAAAGCGGGGCAAUCAAGCGCUAAACCCGCUAAUUGCCCACAUUCGACA-GUUUUGAUAAACU-GUCUUGGAACG-AACGCGU-UGCCAUGGAUAUGACGCAUUUC ........((((((((.....(((((((..((((.......)))).)))))))...(((((((-((((....)))))-))).(((((((-....)))-).))).)))))))))))..... ( -42.40, z-score = -3.45, R) >consensus CUUAUGGGGCGUCGUAAAGCGGGGCAAUCAAGCGCUAAACCCGCUAAUUGCCCACAUUCGACA_GUUUUGAUAAACU_GUCUUGGAACA_AACGCGU_UGCCAUGGAUAUGACGCAUUUC ........((((((((.((((((................))))))..............(((..((((....))))..)))..........................))))))))..... (-18.71 = -19.03 + 0.32)



| Location | 18,405,110 – 18,405,227 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.36 |
| Shannon entropy | 0.31879 |
| G+C content | 0.47060 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -17.60 |
| Energy contribution | -19.70 |
| Covariance contribution | 2.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18405110 117 - 22422827 GAAAUGCGUCAUAUCCAUGGCA-AUGCGUU-UGUUCCAAGACAAGUUUAUCAAAAC-UGUCGAAUGUGGGCAAUUAGCGGGUUUAGCGCUUGAUUGCCCCACUUUACGACGCCCCAUAAG .....(((((.......(((.(-((.....-.)))))).((((.((((....))))-))))...((.((((((((((((.......))).)))))))))))......)))))........ ( -34.10, z-score = -1.87, R) >droEre2.scaffold_4690 8722314 94 - 18748788 GAAAUGCGUCAUACCCAUGGCA-ACGCGUUCGUUUGCAAAAG-AGUUUAUCAAAACUUGUCGAAUGUCGGCAAUUAG------------------------CUUUACGACGGCCCAUAAG .....(((((((....))))).-...(((.(((..((....(-(((((....))))))((((.....)))).....)------------------------)...))))))))....... ( -21.00, z-score = -0.33, R) >droYak2.chrX 17036162 118 - 21770863 GAAAUGCGUCAUAUCCACGUUAUAUUCGUUAUACUCCAAGAC-AGUUUAUCAAAAC-UGUCGAAUGCCGGCAAUUAGCGGGUUUAGCGCCUAAUUGCCCCGCUUUACGACGCCCCAUAAG .....(((((.............................(((-(((((....))))-))))....((.(((((((((.(.(.....).))))))))))..)).....)))))........ ( -33.20, z-score = -3.29, R) >droSec1.super_8 711306 115 - 3762037 GAAAUGCGUCAUAUCCAGGCA--ACGCGUU-CGUUCCAAGAC-AGUUUAUCAAAAC-UGUCGAAUGUGGGCAAUUAGCGGGUUUAGCGCUUGAUUGCCCCGCUUUACGACGCCCCAUAAG .....(((((.......(...--.)(((..-(((((...(((-(((((....))))-))))))))).((((((((((((.......))).)))))))))))).....)))))........ ( -41.50, z-score = -3.78, R) >droSim1.chrX 14210672 116 - 17042790 GAAAUGCGUCAUAUCCAUGGCA-ACGCGUU-CGUUCCAAGAC-AGUUUAUCAAAAC-UGUCGAAUGUGGGCAAUUAGCGGGUUUAGCGCUUGAUUGCCCCGCUUUACGACGCCCCAUAAG .....(((((........(...-.)(((..-(((((...(((-(((((....))))-))))))))).((((((((((((.......))).)))))))))))).....)))))........ ( -41.30, z-score = -3.59, R) >consensus GAAAUGCGUCAUAUCCAUGGCA_ACGCGUU_CGUUCCAAGAC_AGUUUAUCAAAAC_UGUCGAAUGUGGGCAAUUAGCGGGUUUAGCGCUUGAUUGCCCCGCUUUACGACGCCCCAUAAG ..(((((((..............)))))))............................((((((.((((((((((((((.......))).)))))).))))).)).)))).......... (-17.60 = -19.70 + 2.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:05 2011