
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,830,573 – 11,830,666 |
| Length | 93 |
| Max. P | 0.724801 |

| Location | 11,830,573 – 11,830,666 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.65011 |
| G+C content | 0.62758 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.94 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
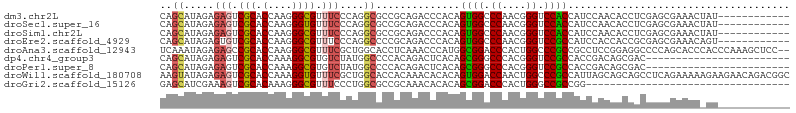
>dm3.chr2L 11830573 93 - 23011544 CAGCAUAGAGAGUCGCACCAAGGGCGUUUCCCAGGCGCCGCAGACCCACAGUGGCCCAACGGGUCCACCAUCCAACACCUCGAGCGAAACUAU------------ ........((..((((......((((((.....))))))...........(((((((...))).))))...............))))..))..------------ ( -25.20, z-score = -0.77, R) >droSec1.super_16 33749 93 - 1878335 CAGCAUAGAGAGUCGCACCAAGGGUGUUUCCCAGGCGCCGCAGACCCACAGUGGCCCAACGGGUCCACCAUCCAACACCUCGAGCGAAACUAU------------ ........((..((((.(...(((((((.....((.(((((.........)))))))...((((.....))))))))))).).))))..))..------------ ( -27.40, z-score = -1.40, R) >droSim1.chr2L 11640675 93 - 22036055 CAGCAUAGAGAGUCGCACCAAGGGCGUUUCCCAGGCGCCGCAGACCCACAGUGGCCCAACGGGUCCACCAUCCAACACCUCGAGCGAAACUAU------------ ........((..((((......((((((.....))))))...........(((((((...))).))))...............))))..))..------------ ( -25.20, z-score = -0.77, R) >droEre2.scaffold_4929 13056483 93 + 26641161 CAGCAUAGAGUGUCGCACCAAGGGCGUUUCCCAGGCCCCGCAGACCCACAGUGGCCCAACGGGUCCGCCAUCCACCACCGCGAGCGAAACAGU------------ .........((.((((.....((((.(.....).))))(((.(.....).(((((((...))).))))...........))).)))).))...------------ ( -25.10, z-score = 0.30, R) >droAna3.scaffold_12943 2600869 103 + 5039921 UCAAAUAGAGAGCCGCACCAAGGGCGUUUCGCUGGCACCUCAAACCCAUGGCGGACCCACUGGCCCGCCGCCUCCGGAGGCCCCAGCACCCACCCAAAGCUCC-- .........(((((((.(....))))....(((((..((((.......((((((.((....)).))))))......))))..)))))...........)))).-- ( -37.52, z-score = -1.85, R) >dp4.chr4_group3 9151318 81 - 11692001 CAGCAUAGAGAGUCGCACCAAAGGCGUGUCUAUGGCCCCACAGACUCACAGCGGGCCCACGGGUCCGCCACCGACAGCGAC------------------------ ...........(((((.......(.(.((((.((....)).)))).).).(((((((....)))))))........)))))------------------------ ( -29.40, z-score = -1.83, R) >droPer1.super_8 339220 81 - 3966273 CAGCAUAGAGAGUCGCACCAAAGGCGUGUCUAUGGCCCCACAGACUCACAGCGGGCCCACGGGUCCGCCACCGACAGCGAC------------------------ ...........(((((.......(.(.((((.((....)).)))).).).(((((((....)))))))........)))))------------------------ ( -29.40, z-score = -1.83, R) >droWil1.scaffold_180708 2480772 105 - 12563649 AAGUAUAGAGAGUCGCACCAAAGGUGUUUCGCUGGCACCACAAACACACAGUGGACCAACUGGCCCGCCAUUAGCAGCAGCCUCAGAAAAAGAAGAACAGACGGC .......(((.((.((......((((((.....))))))...........((((.((....)).))))........)).)))))..................... ( -23.00, z-score = 0.31, R) >droGri2.scaffold_15126 6496110 71 - 8399593 GAGCAUCGAAAGUCGCACAAAGGGCGUUUCCCUGGCGCCGCAAACACACAGCGGACCCACUGGGCCGCCGG---------------------------------- ..((..(....)..))......((((...((((((.(((((.........)))).))))..))).))))..---------------------------------- ( -25.00, z-score = -0.58, R) >consensus CAGCAUAGAGAGUCGCACCAAGGGCGUUUCCCUGGCGCCGCAGACCCACAGUGGCCCAACGGGUCCGCCAUCCACCACCGCGAGCGAAAC_AU____________ ..((.....(((.(((.(....)))).)))....))..............((((.((....)).))))..................................... (-13.00 = -12.94 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:45 2011