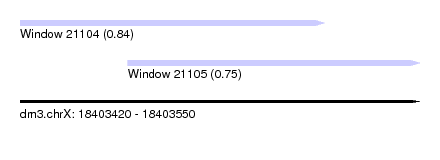
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,403,420 – 18,403,550 |
| Length | 130 |
| Max. P | 0.844333 |

| Location | 18,403,420 – 18,403,519 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 64.60 |
| Shannon entropy | 0.63974 |
| G+C content | 0.56463 |
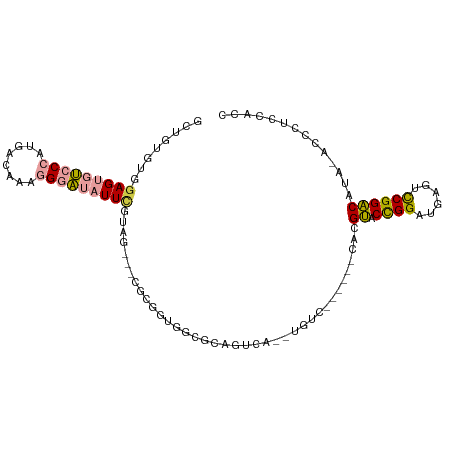
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -10.57 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
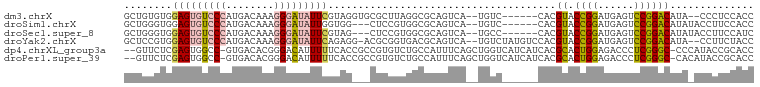
>dm3.chrX 18403420 99 + 22422827 UCCACAAUCUACGGCCGCAGUUGAGUGAUUGCCCCGCUGUGUGGAGUGUCCCAUGACAAAGGGAUAUUCGUAGGUGCGCUUAGGCGCAGUCA--UGUCCAC------ ((((((.....(((..((((((....)))))).)))...))))))(((((((........))))))).(((.(.(((((....))))).).)--)).....------ ( -34.90, z-score = -1.36, R) >droSim1.chrX 14209040 93 + 17042790 CCCACAAUCUGUGGCCGG---UGAGUGAUUGCCCCGCUGGGUGGAGUGUCCCAUGACAAAGGGAUAUUGGUGGCUCCGU---GGCGCAGUCA--UGUCCAC------ .((((.....))))..((---...((((((((.((((.((((.(.(((((((........)))))))...).)))).))---)).)))))))--)..))..------ ( -40.50, z-score = -1.99, R) >droSec1.super_8 709632 93 + 3762037 CCCACAAUCUGUGGCCGG---UGAGUGAUUGCCCCGCUGGGUGGAGUGUCCCAUGACAAAGGGAUAUUCGUAGCUCCGU---GGCGCAGUCA--UGCCCAC------ .((((.....))))..((---.(.((((((((.((((.((((.(((((((((........)))))))))...)))).))---)).)))))))--).)))..------ ( -43.40, z-score = -3.49, R) >droYak2.chrX 17034453 98 + 21770863 ----CUAUCUGUGGCCAAG--UGAGUGAUUGCCCCGCUCCGUGGAGUGUCCCAUGACAAAGGGAUAUUCAGAGG-ACGCGGUGACGCAGUCA--UGUCUAUGUCCAC ----......((((.((((--...((((((((((((((((...(((((((((........)))))))))...))-).)))).)..)))))))--)..)).)).)))) ( -42.40, z-score = -3.87, R) >dp4.chrXL_group3a 342118 104 + 2690836 GCCGCUAUCUGGGAUCGUGGAUAAAUGAGUGUCCAGUUC--UCGAGUGGCC-GUGACACGGGACAUUUUUCACCGCCGUGUCUGCCAUUUCAGCUGGUCAUCAUCAC .((((.(((...))).))))....((((.((.((((((.--..(((((((.-..(((((((..............))))))).))))))).)))))).))))))... ( -36.54, z-score = -1.62, R) >droPer1.super_39 356588 104 - 745454 GCCGCUAUCUGGGAUCGUGGAUAAAUGAGUGUCCAGUUC--UCGAGUGGCC-GUGACACGGGACAUUUUUCACCGCCGUGUCUGCCAUUUCAGCUGGUCAUCAUCAC .((((.(((...))).))))....((((.((.((((((.--..(((((((.-..(((((((..............))))))).))))))).)))))).))))))... ( -36.54, z-score = -1.62, R) >consensus CCCACAAUCUGUGGCCGGG__UGAGUGAUUGCCCCGCUC_GUGGAGUGUCCCAUGACAAAGGGAUAUUCGUAGCUCCGUGU_GGCGCAGUCA__UGUCCAC______ ............((.((........(((((((.((((......(((((((((........)))))))))........))...)).)))))))..)).))........ (-10.57 = -10.57 + 0.01)



| Location | 18,403,455 – 18,403,550 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 62.91 |
| Shannon entropy | 0.65674 |
| G+C content | 0.57579 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -13.66 |
| Energy contribution | -13.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18403455 95 + 22422827 GCUGUGUGGAGUGUCCCAUGACAAAGGGAUAUUCGUAGGUGCGCUUAGGCGCAGUCA--UGUC------CACGUACCGGAUGAGUCCGGACAUA--CCCUCCACC .....((((((((((((........))))))......(((((((....)))).....--....------...((.(((((....)))))))..)--)))))))). ( -35.40, z-score = -1.69, R) >droSim1.chrX 14209072 94 + 17042790 GCUGGGUGGAGUGUCCCAUGACAAAGGGAUAUUGGUGG---CUCCGUGGCGCAGUCA--UGUC------CACGUACCGGAUGAGUCCGGACAUAUACCUUCCACC ....(((((((((((((........))))))).((((.---...(((((.(((....--))))------))))..(((((....))))).....)))).)))))) ( -38.20, z-score = -1.69, R) >droSec1.super_8 709664 94 + 3762037 GCUGGGUGGAGUGUCCCAUGACAAAGGGAUAUUCGUAG---CUCCGUGGCGCAGUCA--UGCC------CACGUACCGGAUGAGUCCGGACAUAUACCUUCCAUC (..((((((((((((((........)))))))))((..---...(((((.(((....--))))------))))..(((((....)))))))...)))))..)... ( -38.30, z-score = -2.61, R) >droYak2.chrX 17034482 100 + 21770863 GCUCCGUGGAGUGUCCCAUGACAAAGGGAUAUUCAGAGG-ACGCGGUGACGCAGUCA--UGUCUAUGUCCACGUACCGGAUGAGUCCGGACAUA--CCUUCUACC ....(((((((((((((........)))))))))(((.(-(((((....))).))).--..))).....))))..(((((....))))).....--......... ( -37.50, z-score = -1.94, R) >dp4.chrXL_group3a 342153 101 + 2690836 --GUUCUCGAGUGGCC-GUGACACGGGACAUUUUUCACCGCCGUGUCUGCCAUUUCAGCUGGUCAUCAUCACGCACUGGAGACCCUCGGGC-CCCAUACCGCACC --..((((.((((((.-..(((((((..............))))))).)))......((((((....)))).))))).))))....(((..-......))).... ( -29.94, z-score = -0.04, R) >droPer1.super_39 356623 101 - 745454 --GUUCUCGAGUGGCC-GUGACACGGGACAUUUUUCACCGCCGUGUCUGCCAUUUCAGCUGGUCAUCAUCACGCACUGGAGACCCUCGGGC-CACAUACCGCACC --((.((((((((((.-..(((((((..............))))))).))))........((((..((........))..)))))))))).-.)).......... ( -29.94, z-score = -0.18, R) >consensus GCUGUGUGGAGUGUCCCAUGACAAAGGGAUAUUCGUAG___CGCGGUGGCGCAGUCA__UGUC______CACGUACCGGAUGAGUCCGGACAUA_ACCCUCCACC ........(((((((((........)))))))))......................................((.((((......)))))).............. (-13.66 = -13.67 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:57:03 2011