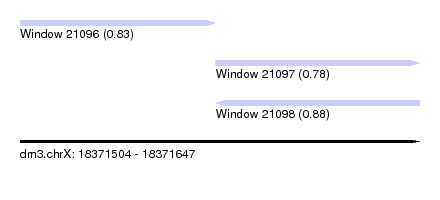
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,371,504 – 18,371,647 |
| Length | 143 |
| Max. P | 0.880217 |

| Location | 18,371,504 – 18,371,574 |
|---|---|
| Length | 70 |
| Sequences | 4 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 86.91 |
| Shannon entropy | 0.20686 |
| G+C content | 0.50046 |
| Mean single sequence MFE | -15.50 |
| Consensus MFE | -12.94 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
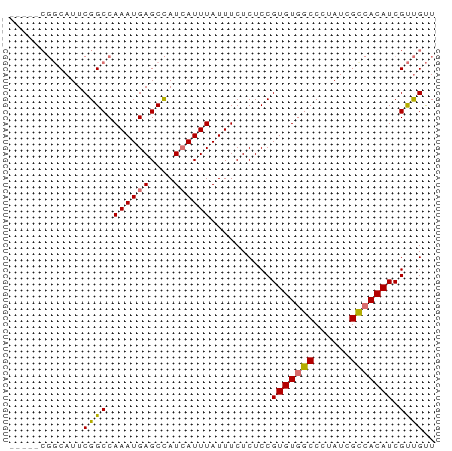
>dm3.chrX 18371504 70 + 22422827 CCAUUCGGCAUUCGGCCAAAUGAGCCAUCAUUUAUUUCUCUCCGUGUGGCCGUAUCGUCACAUCGUUGUU ......(((.....)))((((((....)))))).........((((((((......)))))).))..... ( -15.50, z-score = -1.42, R) >droSim1.chrX 14162842 65 + 17042790 -----CGGCAUUCGGCCAAAUGAGCCAUCAUUUAUUUCUCUCCGUGUUGCCCUAUCGCCACAUCGUUGUU -----.(((.....)))((((((....)))))).........(((((.((......)).))).))..... ( -11.70, z-score = -0.57, R) >droSec1.super_8 677462 65 + 3762037 -----CGGCAUUCGGCCAAAUGAGCCAUCAUUUAUUUCUCUCCGUGUGGCCCUAUCGCCACAUCGUUGUU -----.(((.....)))((((((....)))))).........((((((((......)))))).))..... ( -18.20, z-score = -2.45, R) >droEre2.scaffold_4690 8687762 62 + 18748788 -------CCAUUCGGCCAAAUGAGCUAUAAUUUAUUUCUCUCCGUGUGGCC-CAUCGCCACAUCGCCGCU -------.....((((.....(((..(((...)))..)))...(((((((.-....))))))).)))).. ( -16.60, z-score = -2.74, R) >consensus _____CGGCAUUCGGCCAAAUGAGCCAUCAUUUAUUUCUCUCCGUGUGGCCCUAUCGCCACAUCGUUGUU ............((((.((((((....))))))..........(((((((......))))))).)))).. (-12.94 = -12.88 + -0.06)


| Location | 18,371,574 – 18,371,647 |
|---|---|
| Length | 73 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 70.75 |
| Shannon entropy | 0.51857 |
| G+C content | 0.56981 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -14.78 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
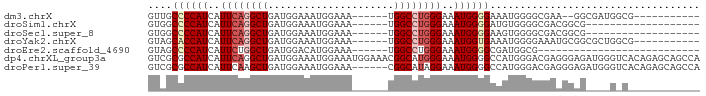
>dm3.chrX 18371574 73 + 22422827 GUUGCCCCAUCAUUCAGGCUGAUGGAAAUGGAAA------UGGCCUGGGAAAUGGGGAAAUGGGGCGAA--GGCGAUGGCG----------- (((.((((((..((((((((..............------.))))))))..)))))).......((...--.))...))).----------- ( -24.96, z-score = -1.85, R) >droSim1.chrX 14162907 67 + 17042790 GUGGCCCCAUCAUUCAGGCUGAUGGAAAUGGAAA------UGGCCUGGGAAAUGGGGAUGUGGGGCGACGGCG------------------- ((.((((((.(((((...((.(.((..((....)------)..))).))......))))))))))).))....------------------- ( -25.80, z-score = -2.32, R) >droSec1.super_8 677527 67 + 3762037 GUGGCCCCAUCAUUCAGGCUGAUGGAAAUGGAAA------UGGCCUGGGAAAUGGGGAAGUGGGGCGACGGCG------------------- ((.(((((((..((((..((.(.((..((....)------)..))).))...))))...))))))).))....------------------- ( -25.20, z-score = -2.21, R) >droYak2.chrX 17002430 75 + 21770863 GUAGCACCAUCAUUCAGGCUGAUGGAAAUGGAAA------UGGCCUGGGAAAUGGUGAAAUGGGGAAAUGCGGCGCUGGCG----------- ....((((((..((((((((..............------.))))))))..))))))...............((....)).----------- ( -19.66, z-score = -0.56, R) >droEre2.scaffold_4690 8687824 59 + 18748788 GUAGCCCCAUCAUUCUGGCUGAUGGACAUGGAAA------UGGCCUGGGAAAUGGGGCGAUGGCG--------------------------- ((.(((((((..(((.(((.......(((....)------))))).)))..)))))))....)).--------------------------- ( -20.81, z-score = -1.27, R) >dp4.chrXL_group3a 318868 92 + 2690836 GUCGCGCCAUCAUUCAGGCUGAUGGAAAUGGAAAUGGAAACGGCAUGGGAAAUGGGGCCAUGGGACGAGGGAGAUGGGUCACAGAGCAGCCA ((.((.(((((.(((.(.((.((((.........((....)).(((.....)))...)))).)).)...)))))))))).)).......... ( -21.30, z-score = 1.06, R) >droPer1.super_39 333326 86 - 745454 GUCGCGCCAUCAUUCAAGCUGAUGGAAAUGGAAA------CGGCAUAGGAAAUGGGGCCAUGGGACGAGGGAGAUGGGUCACAGAGCAGCCA ((.((.(((((.(((..(((.((((...((....------)).(((.....)))...)))).)).)...)))))))))).)).......... ( -20.80, z-score = 0.60, R) >consensus GUAGCCCCAUCAUUCAGGCUGAUGGAAAUGGAAA______UGGCCUGGGAAAUGGGGAAAUGGGGCGACGGAG____G_C____________ ....((((((..((((((((.....................))))))))..))))))................................... (-14.78 = -15.47 + 0.69)

| Location | 18,371,574 – 18,371,647 |
|---|---|
| Length | 73 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 70.75 |
| Shannon entropy | 0.51857 |
| G+C content | 0.56981 |
| Mean single sequence MFE | -16.70 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.880217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
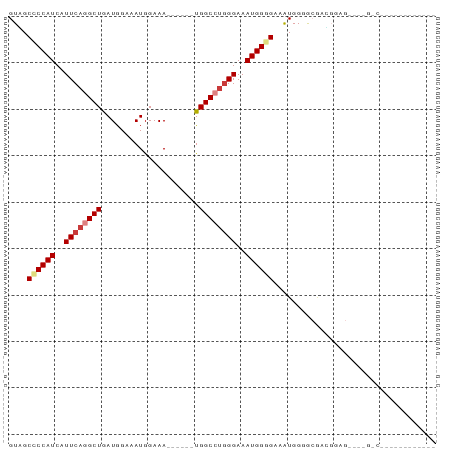
>dm3.chrX 18371574 73 - 22422827 -----------CGCCAUCGCC--UUCGCCCCAUUUCCCCAUUUCCCAGGCCA------UUUCCAUUUCCAUCAGCCUGAAUGAUGGGGCAAC -----------..........--...(((((((.....((((...(((((..------...............))))))))))))))))... ( -19.23, z-score = -1.98, R) >droSim1.chrX 14162907 67 - 17042790 -------------------CGCCGUCGCCCCACAUCCCCAUUUCCCAGGCCA------UUUCCAUUUCCAUCAGCCUGAAUGAUGGGGCCAC -------------------................(((((((...(((((..------...............)))))...))))))).... ( -17.73, z-score = -1.64, R) >droSec1.super_8 677527 67 - 3762037 -------------------CGCCGUCGCCCCACUUCCCCAUUUCCCAGGCCA------UUUCCAUUUCCAUCAGCCUGAAUGAUGGGGCCAC -------------------................(((((((...(((((..------...............)))))...))))))).... ( -17.73, z-score = -1.72, R) >droYak2.chrX 17002430 75 - 21770863 -----------CGCCAGCGCCGCAUUUCCCCAUUUCACCAUUUCCCAGGCCA------UUUCCAUUUCCAUCAGCCUGAAUGAUGGUGCUAC -----------....(((((((((((...................(((((..------...............))))))))).))))))).. ( -16.13, z-score = -1.95, R) >droEre2.scaffold_4690 8687824 59 - 18748788 ---------------------------CGCCAUCGCCCCAUUUCCCAGGCCA------UUUCCAUGUCCAUCAGCCAGAAUGAUGGGGCUAC ---------------------------.......((((((((.....(((..------...............))).....))))))))... ( -17.43, z-score = -1.64, R) >dp4.chrXL_group3a 318868 92 - 2690836 UGGCUGCUCUGUGACCCAUCUCCCUCGUCCCAUGGCCCCAUUUCCCAUGCCGUUUCCAUUUCCAUUUCCAUCAGCCUGAAUGAUGGCGCGAC .(((.(((.((.(((...........))).)).)))............)))................((((((.......))))))...... ( -14.32, z-score = 1.47, R) >droPer1.super_39 333326 86 + 745454 UGGCUGCUCUGUGACCCAUCUCCCUCGUCCCAUGGCCCCAUUUCCUAUGCCG------UUUCCAUUUCCAUCAGCUUGAAUGAUGGCGCGAC .(((.(((.((.(((...........))).)).)))............))).------.........((((((.......))))))...... ( -14.32, z-score = 1.19, R) >consensus ____________G_C____CCCCCUCGCCCCAUUUCCCCAUUUCCCAGGCCA______UUUCCAUUUCCAUCAGCCUGAAUGAUGGGGCCAC ...................................(((((((...(((((.......................)))))...))))))).... (-12.40 = -12.83 + 0.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:57 2011