| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,367,446 – 18,367,536 |
| Length | 90 |
| Max. P | 0.988245 |

| Location | 18,367,446 – 18,367,536 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 55.00 |
| Shannon entropy | 0.80240 |
| G+C content | 0.53794 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -7.62 |
| Energy contribution | -9.95 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.717607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
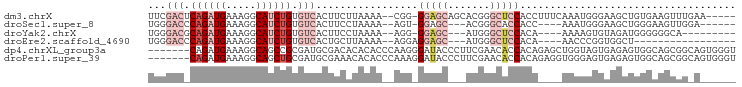
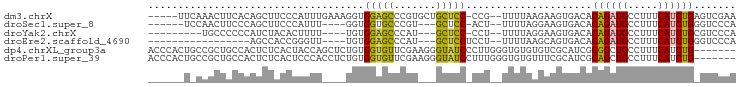
>dm3.chrX 18367446 90 + 22422827 UUCGACUCAGAUGAAAGGCAUCUGUGUCACUUCUUAAAA--CGG-GGAGCAGCACGGGCUCCACCUUUCAAAUGGGAAGCUGUGAAGUUUGAA----- ...(((.((((((.....)))))).)))(((((.....(--(((-(((((.......)))))..(((((.....)))))))))))))).....----- ( -28.70, z-score = -1.65, R) >droSec1.super_8 673479 82 + 3762037 UGGGACCCAGAUGAAAGGCAUCUGUGUCACUUCCUAAAA--AGU-GGAGC---ACGGGCACCACC----AAAUGGGAAGCUGGGAAGUUGGA------ .....(((((......((((....)))).(((((((...--.((-((.((---....)).)))).----...))))))))))))........------ ( -28.20, z-score = -1.51, R) >droYak2.chrX 16988681 79 + 21770863 UGGGACGCAGAUGAAAGGCAUCUGUGUCACUUCCUAAAA--AGG-GGAGC---AUGGGCUCCACA----AAAAGUGUAGAUGGGGGGCA--------- ...((((((((((.....)))))))))).(((((((...--...-(((((---....)))))(((----.....)))...)))))))..--------- ( -31.30, z-score = -4.11, R) >droEre2.scaffold_4690 8684006 72 + 18748788 UGGGACCCAGAUGAAAGGCAUCUGUGUCACUGCUUAAAA--AGGAGGAGC---AUGGGCUCCACA----AACCCGGUGGCU----------------- .......((((((.....)))))).((((((((((....--))).(((((---....)))))...----....))))))).----------------- ( -23.30, z-score = -0.75, R) >dp4.chrXL_group3a 315405 91 + 2690836 -------CAGAUGAAAGGCAGCCGCGAUGCGACACACACCCAAGGGAUACCCUUCGAACACCACAGAGCUGGUAGUGAGAGUGGCAGCGGCAGUGGGU -------..........((.(((((..(((.((.(.((((.(((((...))))).)...((((......)))).))).).)).)))))))).)).... ( -26.90, z-score = -0.29, R) >droPer1.super_39 329911 91 - 745454 -------CAGAUGAAAGGCAGCUGCGAUGCGAAACACACCCAAAGGAUACCCUUCGAACACCACAGAGGUGGGAGUGAGAGUGGCAGCGGCAGUGGGU -------..........((.(((((.((.(....(((.(((.((((....)))).....(((.....)))))).))).).)).))))).))....... ( -26.40, z-score = -0.72, R) >consensus UGGGAC_CAGAUGAAAGGCAUCUGUGUCACUUCAUAAAA__AGG_GGAGC___ACGGGCACCACA____AAAUGGGAAGAUGGGAAGCGG_A______ ...(((.((((((.....)))))).))).................(((((.......))))).................................... ( -7.62 = -9.95 + 2.33)
| Location | 18,367,446 – 18,367,536 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 55.00 |
| Shannon entropy | 0.80240 |
| G+C content | 0.53794 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -9.07 |
| Energy contribution | -9.43 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
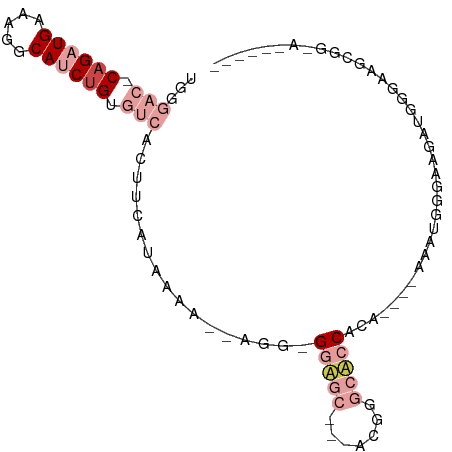
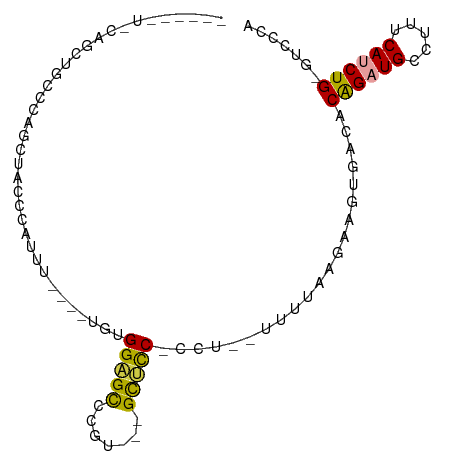
>dm3.chrX 18367446 90 - 22422827 -----UUCAAACUUCACAGCUUCCCAUUUGAAAGGUGGAGCCCGUGCUGCUCC-CCG--UUUUAAGAAGUGACACAGAUGCCUUUCAUCUGAGUCGAA -----........((...(((((....((((((((.(((((.......)))))-)).--)))))))))))(((.((((((.....)))))).))))). ( -27.40, z-score = -2.77, R) >droSec1.super_8 673479 82 - 3762037 ------UCCAACUUCCCAGCUUCCCAUUU----GGUGGUGCCCGU---GCUCC-ACU--UUUUAGGAAGUGACACAGAUGCCUUUCAUCUGGGUCCCA ------............((((((.....----(((((.((....---)).))-)))--.....))))))(((.((((((.....)))))).)))... ( -27.50, z-score = -2.61, R) >droYak2.chrX 16988681 79 - 21770863 ---------UGCCCCCCAUCUACACUUUU----UGUGGAGCCCAU---GCUCC-CCU--UUUUAGGAAGUGACACAGAUGCCUUUCAUCUGCGUCCCA ---------.............(((((((----((.(((((....---)))))-...--...)))))))))((.((((((.....)))))).)).... ( -21.30, z-score = -3.20, R) >droEre2.scaffold_4690 8684006 72 - 18748788 -----------------AGCCACCGGGUU----UGUGGAGCCCAU---GCUCCUCCU--UUUUAAGCAGUGACACAGAUGCCUUUCAUCUGGGUCCCA -----------------.((....(((..----...(((((....---))))).)))--......)).(.(((.((((((.....)))))).))).). ( -20.60, z-score = -0.47, R) >dp4.chrXL_group3a 315405 91 - 2690836 ACCCACUGCCGCUGCCACUCUCACUACCAGCUCUGUGGUGUUCGAAGGGUAUCCCUUGGGUGUGUGUCGCAUCGCGGCUGCCUUUCAUCUG------- .......((((((((.((.(.((((((((......)))))..(.(((((...))))).)))).).)).)))..))))).............------- ( -28.40, z-score = -0.70, R) >droPer1.super_39 329911 91 + 745454 ACCCACUGCCGCUGCCACUCUCACUCCCACCUCUGUGGUGUUCGAAGGGUAUCCUUUGGGUGUGUUUCGCAUCGCAGCUGCCUUUCAUCUG------- .......((.(((((............((((.....))))..((((((....))))))((((((...))))))))))).))..........------- ( -26.70, z-score = -1.64, R) >consensus ______U_CAGCUGCCCAGCUACCCAUUU____UGUGGAGCCCGU___GCUCC_CCU__UUUUAAGAAGUGACACAGAUGCCUUUCAUCUG_GUCCCA ....................................(((((.......))))).................(((.((((((.....)))))).)))... ( -9.07 = -9.43 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:55 2011