| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,349,053 – 18,349,148 |
| Length | 95 |
| Max. P | 0.936757 |

| Location | 18,349,053 – 18,349,148 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.30 |
| Shannon entropy | 0.66060 |
| G+C content | 0.44451 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -11.95 |
| Energy contribution | -12.53 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
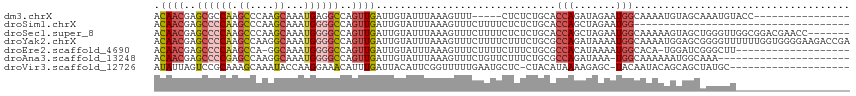
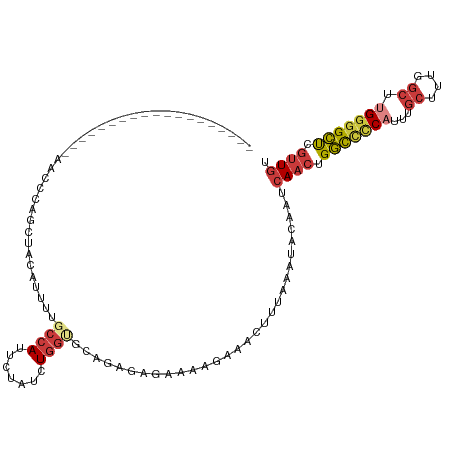
>dm3.chrX 18349053 95 + 22422827 ACAACGAGCGCCAAGCCCAAGCAAAUGAGGCCAGUUGAUUGUAUUUAAAGUUU-----CUCUCUGCACCAGAUAGAAUGGCAAAAUGUAGCAAAUGUACC---------------- .........((((.(((((......)).)))....................((-----((.((((...)))).))))))))...................---------------- ( -15.90, z-score = 0.86, R) >droSim1.chrX 14172737 80 + 17042790 ACAACGAGCCCCAAGCCCAAGCAAAUGGGGCCAGUUGAUUGUAUUUAAAGUUUCUUUUCUCUCUGCACCAGCUAGAAUGG------------------------------------ .((((..((((((.((....))...))))))..))))................((.((((..(((...)))..)))).))------------------------------------ ( -20.20, z-score = -0.97, R) >droSec1.super_8 652225 109 + 3762037 ACAACGAGCCCCAAGCCCAAGCAAAUGGGGCCAGUUGAUUGUAUUUAAAGUUUCUUUUCUCUCUGCACCAGCUAGAAUGGCAAAAAGUAGCUGGGUUGGCGGACGAACC------- .((((..((((((.((....))...))))))..))))......................(((((((.(((((((.............)))))))....))))).))...------- ( -34.42, z-score = -1.33, R) >droYak2.chrX 16968344 116 + 21770863 ACAACGAGCCCCAAGCCAAGGCAAAUGGGGCCAGUUGAUUGUAUUUAAAGUUUCUUUUCUUUCUGCGCCAGAUAAAAUGGCAAAAUGGAGCGGGGUUUUUUGGUGGGGAAGACCGA .((((..((((((.((....))...))))))..))))....................(((((((.(((((((.((..(.((........)).)..)).)))))))))))))).... ( -38.10, z-score = -1.36, R) >droEre2.scaffold_4690 8666252 95 + 18748788 ACAACGAGCCCCAAGCCA-GGCAAAUGGGGCCAGUUGAUUGUAUUUAAAGUUUCUUUUCUUUCUGCGCCACAUAAAAUGGCACA-UGGAUCGGGCUU------------------- .((((..((((((.((..-.))...))))))..))))..........((((((....(((...((.((((.......)))).))-.)))..))))))------------------- ( -25.50, z-score = -0.36, R) >droAna3.scaffold_13248 4017191 93 - 4840945 ACAACGAGCCCCGAGCCAAGGCAAAUGGGGCCAGUUGAUUGUAUUUAAAGUUUCUGUUCUUUCUGCGCCAGAUAAA-UGGCAAAAAAUGGCAAA---------------------- .((((..((((((.((....))...))))))..)))).((((((((((((........))))....((((......-))))...)))).)))).---------------------- ( -26.80, z-score = -1.20, R) >droVir3.scaffold_12726 2145610 94 + 2840439 AUAUUAGUCCGCAAAGCAAAUACCAAGGAAACAUUUGAUUACAUUCGGUUUUUGAAUGCUC-CUACAUAAAAGAGC-UACAAUACAGCAGCUAUGC-------------------- ..........(((.(((........((((..((((..(..((.....))..)..)))).))-))..........((-(.......))).))).)))-------------------- ( -18.10, z-score = -1.46, R) >consensus ACAACGAGCCCCAAGCCAAAGCAAAUGGGGCCAGUUGAUUGUAUUUAAAGUUUCUUUUCUCUCUGCACCAGAUAGAAUGGCAAAAAGUAGCAAGGUU___________________ .((((..((((((.((....))...))))))..))))............................................................................... (-11.95 = -12.53 + 0.57)
| Location | 18,349,053 – 18,349,148 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.30 |
| Shannon entropy | 0.66060 |
| G+C content | 0.44451 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -15.81 |
| Energy contribution | -16.86 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
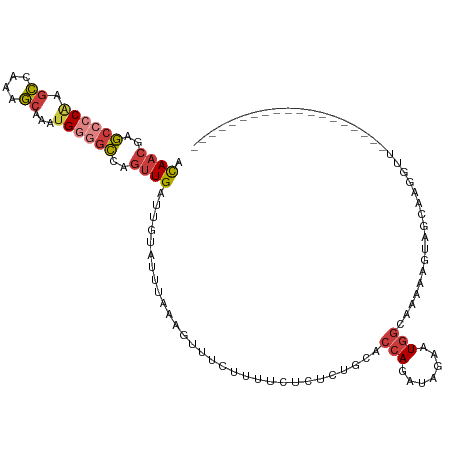
>dm3.chrX 18349053 95 - 22422827 ----------------GGUACAUUUGCUACAUUUUGCCAUUCUAUCUGGUGCAGAGAG-----AAACUUUAAAUACAAUCAACUGGCCUCAUUUGCUUGGGCUUGGCGCUCGUUGU ----------------((((....)))).......((((((((.((((...)))).))-----))...................((((.((......))))))))))......... ( -17.70, z-score = 1.29, R) >droSim1.chrX 14172737 80 - 17042790 ------------------------------------CCAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGU ------------------------------------...((((..(((...)))..))))...................((((.(((((((...((....)).))))))).)))). ( -24.70, z-score = -1.39, R) >droSec1.super_8 652225 109 - 3762037 -------GGUUCGUCCGCCAACCCAGCUACUUUUUGCCAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGU -------..(((.((.((....(((((((.............))))))).)).))..)))...................((((.(((((((...((....)).))))))).)))). ( -31.92, z-score = -0.57, R) >droYak2.chrX 16968344 116 - 21770863 UCGGUCUUCCCCACCAAAAAACCCCGCUCCAUUUUGCCAUUUUAUCUGGCGCAGAAAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCCUUGGCUUGGGGCUCGUUGU ((..(((((..(.(((.........((........)).........))).)..).))))...))...............((((.(((((((...((....)).))))))).)))). ( -27.67, z-score = -0.44, R) >droEre2.scaffold_4690 8666252 95 - 18748788 -------------------AAGCCCGAUCCA-UGUGCCAUUUUAUGUGGCGCAGAAAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCC-UGGCUUGGGGCUCGUUGU -------------------............-((((((((.....))))))))..........................((((.(((((((...((.-..)).))))))).)))). ( -30.00, z-score = -1.92, R) >droAna3.scaffold_13248 4017191 93 + 4840945 ----------------------UUUGCCAUUUUUUGCCA-UUUAUCUGGCGCAGAAAGAACAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCCUUGGCUCGGGGCUCGUUGU ----------------------((((...((((((((((-......))))).)))))...))))...............((((.((((((....((....))..)))))).)))). ( -27.60, z-score = -1.95, R) >droVir3.scaffold_12726 2145610 94 - 2840439 --------------------GCAUAGCUGCUGUAUUGUA-GCUCUUUUAUGUAG-GAGCAUUCAAAAACCGAAUGUAAUCAAAUGUUUCCUUGGUAUUUGCUUUGCGGACUAAUAU --------------------...(((((((.........-((((((......))-))))...((((.(((((..................))))).))))....)))).))).... ( -19.27, z-score = -0.30, R) >consensus ___________________AACCCAGCUACAUUUUGCCAUUCUAUCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUUGGCUUGGGGCUCGUUGU ...................................((((.......)))).............................((((.(((((((...((....)).))))))).)))). (-15.81 = -16.86 + 1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:49 2011