
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,330,388 – 18,330,500 |
| Length | 112 |
| Max. P | 0.804680 |

| Location | 18,330,388 – 18,330,500 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.17 |
| Shannon entropy | 0.69086 |
| G+C content | 0.59220 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.71 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
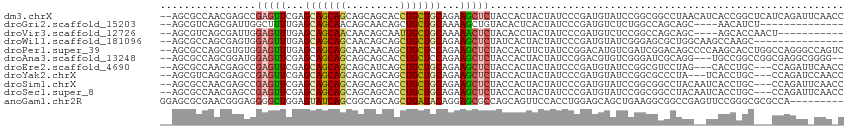
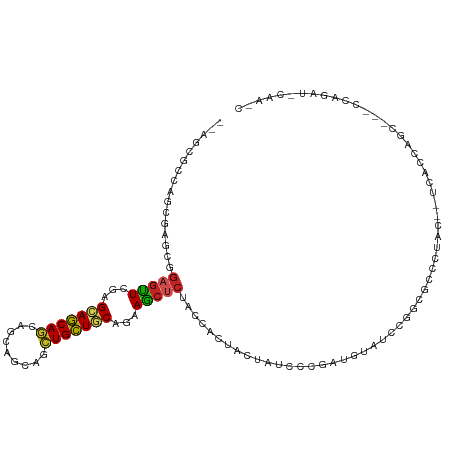
>dm3.chrX 18330388 112 - 22422827 --AGCGCCAACGAGCCGAGUUCGAGCAGCAGCAGCAGCACCUGCUGCAGAAGCUCUACCACUACUAUCCCGAUGUAUCCGGCGGCCUAACAUCACCGGCUCAUCAGAUUCAACC --.........((((((.((..((((..(.(((((((...))))))).)..))))...............(((((..((...))....)))))))))))))............. ( -33.70, z-score = -1.66, R) >droGri2.scaffold_15203 1046467 94 - 11997470 --AGCGUCAGCGAUUGGCUUUUGAGCAGCAACAGCAACAGCUGCUGCAAAAGCUGUACACUCACUAUCCCGAUGUCUCUGGCCAGCAGC----AACAUCU-------------- --.((....))....((((((((((((((..........)))))).))))))))................(((((..(((.....))).----.))))).-------------- ( -29.20, z-score = -0.60, R) >droVir3.scaffold_12726 2119590 97 - 2840439 --AGCGUCAGCGAUUGGAGUUUGAGCAGCAACAACAGCAAUUGCUGCAAAAACUCUACACCUACUAUCCCGAUGUCUCCGGCCAGCAGC----AGCACCAACU----------- --.((....((...((((((((..(((((((.........)))))))..))))))))...........(((.......)))...))...----.)).......----------- ( -25.30, z-score = -0.91, R) >droWil1.scaffold_181096 7008060 97 + 12416693 --AGCGCCAGCGAGUGGAGUUUGAGCAGCAGCAACAGCAGCUGCUGCAGAAGCUCUAUCACUACUAUCCCGAUGUAUCGGGAGCGCUGGCAAGCCAAGC--------------- --...(((((((.(((((((((..((((((((.......))))))))..)))))))))........((((((....)))))).))))))).........--------------- ( -50.20, z-score = -4.88, R) >droPer1.super_39 296746 112 + 745454 --AGCGCCAGCGUGUGGAGUUUGAGCAGCAGCAACAACAGCUGCUCCAGAAGCUCUACCACUUCUAUCCGGACAUGUCGAUCGGACAGCCCCAAGCACCUGGCCAGGGCCAGUC --.......((((((((((((((((((((..........)))))))...)))))))).)))........((...((((.....))))...))..))..(((((....))))).. ( -40.70, z-score = -1.39, R) >droAna3.scaffold_13248 3982226 107 + 4840945 --AGCGCCAGCGGAUGGAGUUCGAGCAGCAGCAGCAGCACCUGCUCCAGAAGCUCUACCACUACUAUCCGGACGUGUCGGGAUCGCAGG---UGCCGGCCGGCGAGGCGGGG-- --.(((((.((((.(((((((.((((((..((....))..))))))....)))))))))......((((.((....)).)))).)).))---)))(.(((.....))).)..-- ( -44.00, z-score = 0.12, R) >droEre2.scaffold_4690 8649592 106 - 18748788 --AGCGCCAACGAGCCGAGUUCGAGCAGCAGCAGCAUCAGCUGCUGCAGAAGCUCUACCACUACUAUCCCGAUGUAUCCGGCGUCCUAG---CACCUGC---CCAGAUUCAACC --.(((((...((((...))))((((.((((((((....))))))))....))))........................)))))....(---(....))---............ ( -31.90, z-score = -1.71, R) >droYak2.chrX 16948650 106 - 21770863 --AGCGUCAGCGAGCCGAGUUCGAGCAGCAGCAGCAGCAGCUGCUGCAGAAGCUCUACCACUACUAUCCCGAUGUAUCCGGCGCCCUA---UCACCUGC---CCAGAUCCAACC --.(((((...((((...))))((((.((((((((....))))))))....))))...............)))))(((.((.((....---......))---)).)))...... ( -31.40, z-score = -0.99, R) >droSim1.chrX 14143774 109 - 17042790 --AGCGCCAACGAGCCGAGUUCGAGCAGCAGCAGCAGCACCUGCUGCAGAAGCUCUACCACUACUAUCCCGAUGUAUCCGGCGGCCUACAAUCACCUGC---CCAGAUUCAACC --..((((...((((...))))((((..(.(((((((...))))))).)..))))........................))))................---............ ( -27.20, z-score = -0.16, R) >droSec1.super_8 634073 109 - 3762037 --AGCGCCAACGAGCCGAGUUCGAGCAGCAGCAGCAGCACCUGCUGCAGAAGCUCUACCACUACUAUCCCGAUGUAUCCGGCGGCCUACAAUCACCUGC---CCAGAUUCAACC --..((((...((((...))))((((..(.(((((((...))))))).)..))))........................))))................---............ ( -27.20, z-score = -0.16, R) >anoGam1.chr2R 29844241 105 + 62725911 GGAGCGCGAACGGGAGGGGCUGGAGUAUCAGCGGCAGCAGCUGAUACAGGAGCGCCAGCAGUUCCACCUGGAGCAGCUGAAGGCGGCCGAGUUCCGGGCGCGCCA--------- .(.(((((..(((((..(((((..((((((((.......))))))))........((((.(((((....))))).))))....)))))...)))))..)))))).--------- ( -53.00, z-score = -2.78, R) >consensus __AGCGCCAGCGAGCGGAGUUCGAGCAGCAGCAGCAGCAGCUGCUGCAGAAGCUCUACCACUACUAUCCCGAUGUAUCCGGCGCCCUAC__UCACCAGC___CCAGAU_CAA_C ................(((((...(((((((.........)))))))...)))))........................................................... (-13.68 = -13.71 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:43 2011