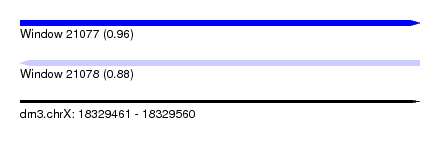
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,329,461 – 18,329,560 |
| Length | 99 |
| Max. P | 0.964213 |

| Location | 18,329,461 – 18,329,560 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.60 |
| Shannon entropy | 0.48479 |
| G+C content | 0.45650 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -16.82 |
| Energy contribution | -16.66 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
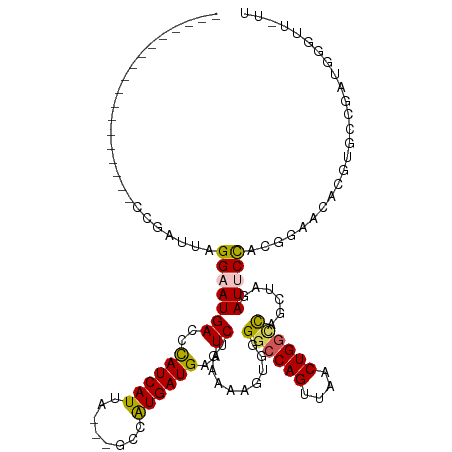
>dm3.chrX 18329461 99 + 22422827 ------------------CCGAUUAGGAAUGACCCAUCAUUA---GCCAUGAUGAAUCUGGUAAGUGGGGCCAGUUAACUGACCAGCUAGAUUCCACGGAGCACGUGCCGAUGGUUUAUU ------------------((.....))(((((.(((((....---........(((((((((...(((...(((....))).))))))))))))((((.....))))..))))).))))) ( -30.00, z-score = -1.60, R) >droSec1.super_8 633174 102 + 3762037 ------------------CCGAUUAGGAAUGACCCAUCAUUAUGAAUCGUGAUGAAUCUCAAAAGUGGGGCCAGUUAACUGGCCAGCUAGAUUCCACGGAACACGUGCCGAUGGGUUGUU ------------------((.....))((..(((((((..((((..(((((..((((((....(((..((((((....)))))).))))))))))))))....))))..)))))))..)) ( -40.30, z-score = -4.16, R) >droSim1.chrX 14142824 102 + 17042790 ------------------CCGAUUAGGAAUGACCCAUCAUUAUGAAUCGUGAUGAAUCUGAAAAGUGGGGCCAGUUAACUGGGCAGCUAGAUUCCACGGAGCACGUGCCGAUGGGUUGUU ------------------((.....))((..(((((((........(((((..((((((....(((..(.((((....)))).).)))))))))))))).((....)).)))))))..)) ( -36.30, z-score = -2.74, R) >droYak2.chrX 16947743 113 + 21770863 GCCAUAAGUAUUUAUACGCCGAUUAGGCAUGACCUAUCAUUA---GCAAUGAUGAAUCUGAAUAAUGGGGCCAGUAAGCUGGUUAACUGGAUUCCAUCAAACACGUGCUGCUGAUU---- ..(((..(((....)))(((.....))))))....((((.((---(((.((((((((((.........((((((....))))))....))))).)))))......))))).)))).---- ( -27.52, z-score = -0.46, R) >droEre2.scaffold_4690 8648841 90 + 18748788 ------------------CCGAGCAGGAAUGACCCAUCAUUA---GCUAUGAUGAAUCUGGCAAAUGUGGCCAGUAAGCUGGUUAACUAGAUGCUGAUGGAAAUAU-UUAUU-------- ------------------((((((..........((((((..---...)))))).((((((......(((((((....))))))).)))))))))..)))......-.....-------- ( -23.00, z-score = -1.30, R) >consensus __________________CCGAUUAGGAAUGACCCAUCAUUA___GCCAUGAUGAAUCUGAAAAGUGGGGCCAGUUAACUGGCCAGCUAGAUUCCACGGAACACGUGCCGAUGGGUU_UU .........................(((((((..((((((........))))))..))..........((((((....))))))......)))))......................... (-16.82 = -16.66 + -0.16)

| Location | 18,329,461 – 18,329,560 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.60 |
| Shannon entropy | 0.48479 |
| G+C content | 0.45650 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.84 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
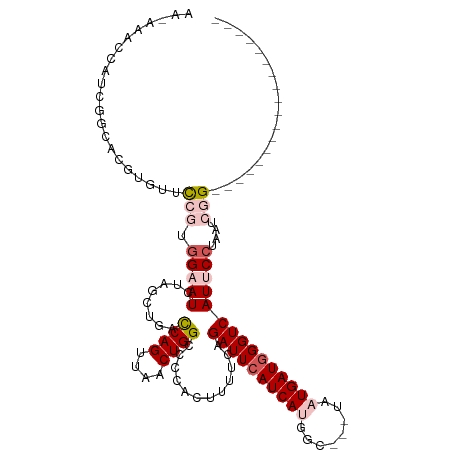
>dm3.chrX 18329461 99 - 22422827 AAUAAACCAUCGGCACGUGCUCCGUGGAAUCUAGCUGGUCAGUUAACUGGCCCCACUUACCAGAUUCAUCAUGGC---UAAUGAUGGGUCAUUCCUAAUCGG------------------ ...........(((....)))(((.(((((......((((((....))))))..........((((((((((...---..)))))))))))))))....)))------------------ ( -29.40, z-score = -1.80, R) >droSec1.super_8 633174 102 - 3762037 AACAACCCAUCGGCACGUGUUCCGUGGAAUCUAGCUGGCCAGUUAACUGGCCCCACUUUUGAGAUUCAUCACGAUUCAUAAUGAUGGGUCAUUCCUAAUCGG------------------ ....((((((((....(((...((((((((((((..((((((....))))))...))....))))))..))))...)))..)))))))).............------------------ ( -33.10, z-score = -2.87, R) >droSim1.chrX 14142824 102 - 17042790 AACAACCCAUCGGCACGUGCUCCGUGGAAUCUAGCUGCCCAGUUAACUGGCCCCACUUUUCAGAUUCAUCACGAUUCAUAAUGAUGGGUCAUUCCUAAUCGG------------------ ....((((((((((....))).((((((((((((.((.((((....))))...))))....))))))..)))).........))))))).............------------------ ( -26.30, z-score = -1.50, R) >droYak2.chrX 16947743 113 - 21770863 ----AAUCAGCAGCACGUGUUUGAUGGAAUCCAGUUAACCAGCUUACUGGCCCCAUUAUUCAGAUUCAUCAUUGC---UAAUGAUAGGUCAUGCCUAAUCGGCGUAUAAAUACUUAUGGC ----.((((..((((..((..((((((...(((((..........)))))..))))))..))((....))..)))---)..))))..((((((((.....)))(((....)))..))))) ( -26.80, z-score = -0.60, R) >droEre2.scaffold_4690 8648841 90 - 18748788 --------AAUAA-AUAUUUCCAUCAGCAUCUAGUUAACCAGCUUACUGGCCACAUUUGCCAGAUUCAUCAUAGC---UAAUGAUGGGUCAUUCCUGCUCGG------------------ --------.....-........................(((((....((((.......))))((((((((((...---..))))))))))......))).))------------------ ( -19.70, z-score = -1.47, R) >consensus AA_AAACCAUCGGCACGUGUUCCGUGGAAUCUAGCUGACCAGUUAACUGGCCCCACUUUUCAGAUUCAUCAUGGC___UAAUGAUGGGUCAUUCCUAAUCGG__________________ .........................(((((........((((....))))............((((((((((........)))))))))))))))......................... (-14.80 = -15.84 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:42 2011