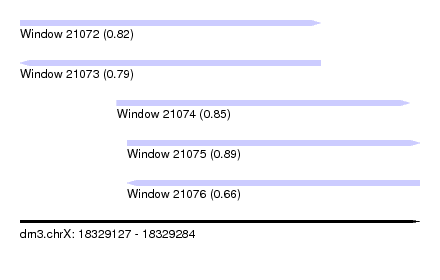
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,329,127 – 18,329,284 |
| Length | 157 |
| Max. P | 0.892667 |

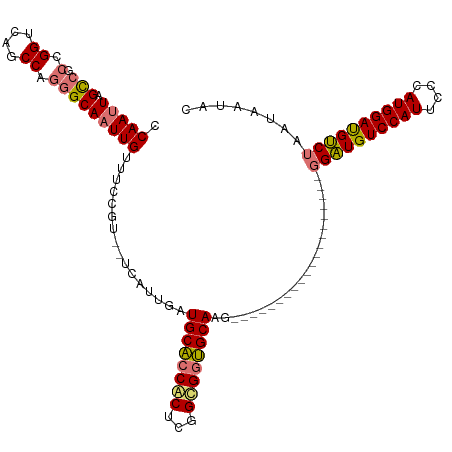
| Location | 18,329,127 – 18,329,245 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Shannon entropy | 0.33922 |
| G+C content | 0.53556 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -27.95 |
| Energy contribution | -28.07 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18329127 118 + 22422827 CCAAUUAGCCGCCGGCCAACCAGGGCAAUUGUCUCCGU--UCAUUGAUGCACCACUCGGCGGUGCAAGUGGUGGUGGACUCGAGCAGAGGAUGUCCAUUCCCAUGGAUGUCUAAUAAUAC .(((((.(((.(.((....)).)))))))))....(((--((.....((((((.......)))))).((((..((((((((........)).))))))..)))))))))........... ( -38.00, z-score = -0.32, R) >droSec1.super_8 632901 98 + 3762037 CCAAUUAGCCGCCGGUCAGCCAGGGCAAUUGUUUCCGU--UCAUUGAUGCACCACUCGGUGGUGCACG--------------------GGAUGUCCAUUCCUAUGGAUGUCUAAUAAUAC .(((((.(((.(.((....)).)))))))))...((((--........(((((((...))))))))))--------------------)(((((((((....)))))))))......... ( -33.80, z-score = -2.08, R) >droSim1.chrX 14142503 98 + 17042790 CCAAUUAGUCGCCGGUCAGCCAGGGCAAUUGUUUCCGU--UCAUUGAUGCACCACUCGGUGGUGCACG--------------------GGAUGUCCAUUCCCAUGGAUGUCUAAUAAUAC ...(((((.((((((....))..))).......(((((--.......((((((((...)))))))).(--------------------(((.......))))))))).).)))))..... ( -31.80, z-score = -1.49, R) >droYak2.chrX 16947434 107 + 21770863 CCAAUUAGCCAGCGGGCAGCCAGUGCACUUGUUUCCAUACCCAUGGAUGCCCCGCGCCGCGUGGCAAGUGG-------------CAAGUGGUGUCCAUUCCCAUGGACGCCUAAUAAUAC .......(((((((((((((....)).......(((((....))))))).)))))(((....)))...)))-------------)....(((((((((....)))))))))......... ( -41.20, z-score = -2.04, R) >consensus CCAAUUAGCCGCCGGUCAGCCAGGGCAAUUGUUUCCGU__UCAUUGAUGCACCACUCGGCGGUGCAAG____________________GGAUGUCCAUUCCCAUGGAUGUCUAAUAAUAC .(((((.(((.(.((....)).)))))))))................((((((((...))))))))......................((((((((((....))))))))))........ (-27.95 = -28.07 + 0.12)

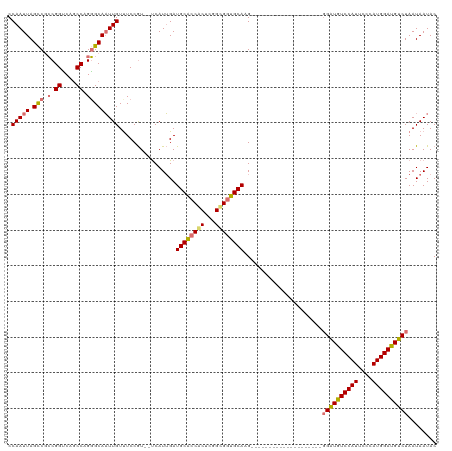
| Location | 18,329,127 – 18,329,245 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Shannon entropy | 0.33922 |
| G+C content | 0.53556 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.786116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18329127 118 - 22422827 GUAUUAUUAGACAUCCAUGGGAAUGGACAUCCUCUGCUCGAGUCCACCACCACUUGCACCGCCGAGUGGUGCAUCAAUGA--ACGGAGACAAUUGCCCUGGUUGGCCGGCGGCUAAUUGG .........((..(((((....)))))..))(((((.(((.......(((((((((......)))))))))......)))--.))))).((((((((((((....)))).))).))))). ( -40.62, z-score = -1.41, R) >droSec1.super_8 632901 98 - 3762037 GUAUUAUUAGACAUCCAUAGGAAUGGACAUCC--------------------CGUGCACCACCGAGUGGUGCAUCAAUGA--ACGGAAACAAUUGCCCUGGCUGACCGGCGGCUAAUUGG ((.(((((.((..(((((....)))))..)).--------------------.(((((((((...))))))))).)))))--)).....((((((((((((....)))).))).))))). ( -32.70, z-score = -2.10, R) >droSim1.chrX 14142503 98 - 17042790 GUAUUAUUAGACAUCCAUGGGAAUGGACAUCC--------------------CGUGCACCACCGAGUGGUGCAUCAAUGA--ACGGAAACAAUUGCCCUGGCUGACCGGCGACUAAUUGG .....(((((...(((..((((.......)))--------------------)(((((((((...)))))))))......--..))).....(((((..((....))))))))))))... ( -29.70, z-score = -1.24, R) >droYak2.chrX 16947434 107 - 21770863 GUAUUAUUAGGCGUCCAUGGGAAUGGACACCACUUG-------------CCACUUGCCACGCGGCGCGGGGCAUCCAUGGGUAUGGAAACAAGUGCACUGGCUGCCCGCUGGCUAAUUGG .........((.((((((....)))))).))....(-------------(((...(((....)))(((((.((.(((((.(..((....))..).)).))).)))))))))))....... ( -43.20, z-score = -1.49, R) >consensus GUAUUAUUAGACAUCCAUGGGAAUGGACAUCC____________________CGUGCACCACCGAGUGGUGCAUCAAUGA__ACGGAAACAAUUGCCCUGGCUGACCGGCGGCUAAUUGG .........((..(((((....)))))..)).......................((((((((...))))))))................((((((((((((....)))).))).))))). (-26.66 = -26.72 + 0.06)

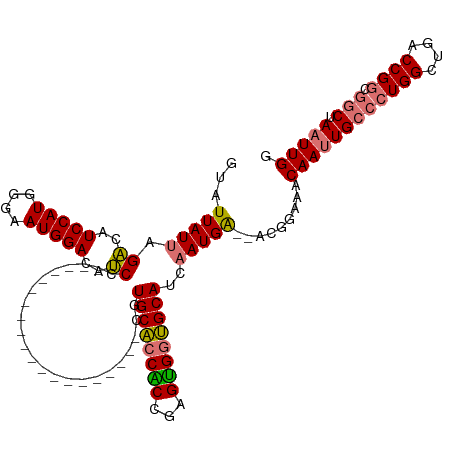
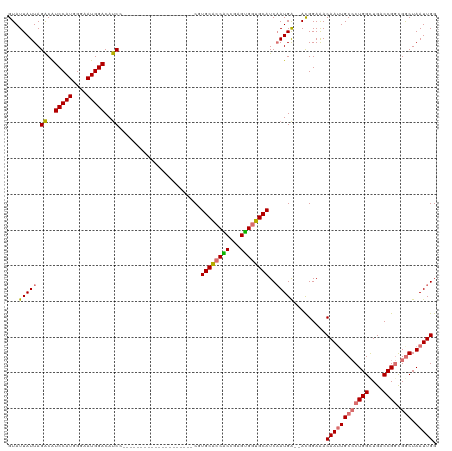
| Location | 18,329,165 – 18,329,280 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.12 |
| Shannon entropy | 0.44141 |
| G+C content | 0.57194 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -25.45 |
| Energy contribution | -26.07 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18329165 115 + 22422827 UCAUUGAUGCACCACUCGGCGGUGCAAGUGGUGGUGGACUCGAGCAGAGGAUGUCCAUUCCCAUGGAUGUCUAAUAAUACCCCAACAAUCGAAGUCCGUCGCCGUUGCGACUGCG ..................(((((((((.(((((..((((((((.....((((((((((....))))))))))................))).)))))..))))))))).))))). ( -41.30, z-score = -1.78, R) >droSec1.super_8 632939 95 + 3762037 UCAUUGAUGCACCACUCGGUGGUGCACG--------------------GGAUGUCCAUUCCUAUGGAUGUCUAAUAAUACGCCCACAAUCGAAGCCCGUCGUGGUCGCGGCUGCG ((((((.((((((((...))))))))..--------------------((((((((((....)))))))))).............)))).)).((..(((((....))))).)). ( -33.40, z-score = -1.31, R) >droSim1.chrX 14142541 95 + 17042790 UCAUUGAUGCACCACUCGGUGGUGCACG--------------------GGAUGUCCAUUCCCAUGGAUGUCUAAUAAUACGCCCACAAUCGAAGCCCGUCGUGGUCGCGGCUGCG ((((((.((((((((...))))))))..--------------------((((((((((....)))))))))).............)))).)).((..(((((....))))).)). ( -32.70, z-score = -0.86, R) >droYak2.chrX 16947474 88 + 21770863 CCAUGGAUGCCCCGCGCCGCGUGGCAAGUGG-------------CAAGUGGUGUCCAUUCCCAUGGACGCCUAAUAAUACACCCGCAAUCGAAGCCCGCCA-------------- .......((((.(((...))).))))..(((-------------(....(((((((((....))))))))).............((.......))..))))-------------- ( -29.00, z-score = -0.64, R) >consensus UCAUUGAUGCACCACUCGGCGGUGCAAG____________________GGAUGUCCAUUCCCAUGGAUGUCUAAUAAUACGCCCACAAUCGAAGCCCGUCGUGGUCGCGGCUGCG ...((((((((((((...))))))).......................((((((((((....))))))))))...............))))).....(((((....))))).... (-25.45 = -26.07 + 0.62)


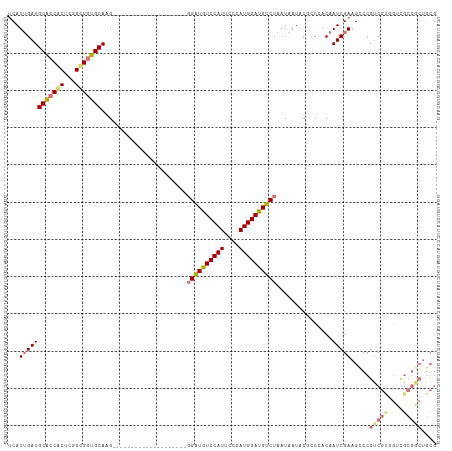
| Location | 18,329,169 – 18,329,284 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Shannon entropy | 0.23956 |
| G+C content | 0.56796 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -32.27 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18329169 115 + 22422827 UGAUGCACCACUCGGCGGUGCAAGUGGUGGUGGACUCGAGCAGAGGAUGUCCAUUCCCAUGGAUGUCUAAUAAUACCCCAACAAUCGAAGUCCGUCGCCGUUGCGACUGCGACUG ...((((((.......))))))...((((..((((((((.....((((((((((....))))))))))................))).)))))..))))(((((....))))).. ( -43.10, z-score = -2.14, R) >droSec1.super_8 632943 95 + 3762037 UGAUGCACCACUCGGUGGUGCACG--------------------GGAUGUCCAUUCCUAUGGAUGUCUAAUAAUACGCCCACAAUCGAAGCCCGUCGUGGUCGCGGCUGCGACUG .((((((((((...)))))))...--------------------((((((((((....))))))))))...............))).......((((..((....))..)))).. ( -35.60, z-score = -1.79, R) >droSim1.chrX 14142545 95 + 17042790 UGAUGCACCACUCGGUGGUGCACG--------------------GGAUGUCCAUUCCCAUGGAUGUCUAAUAAUACGCCCACAAUCGAAGCCCGUCGUGGUCGCGGCUGCGACUG .((((((((((...)))))))...--------------------((((((((((....))))))))))...............))).......((((..((....))..)))).. ( -34.90, z-score = -1.32, R) >consensus UGAUGCACCACUCGGUGGUGCACG____________________GGAUGUCCAUUCCCAUGGAUGUCUAAUAAUACGCCCACAAUCGAAGCCCGUCGUGGUCGCGGCUGCGACUG .((((((((((...))))))).......................((((((((((....))))))))))...............))).......((((((((....)))))))).. (-32.27 = -32.50 + 0.23)

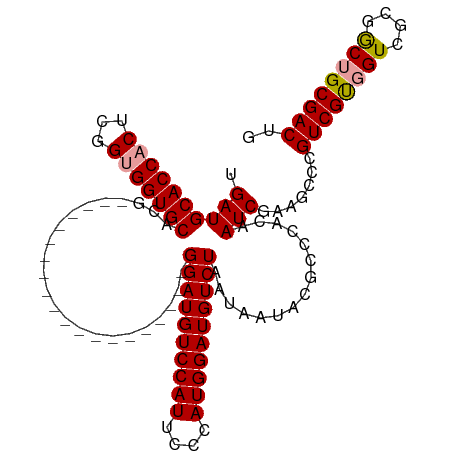
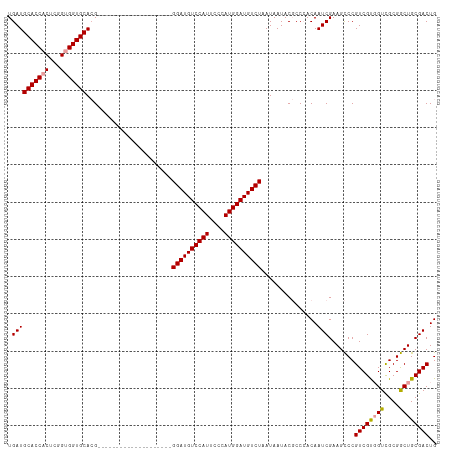
| Location | 18,329,169 – 18,329,284 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Shannon entropy | 0.23956 |
| G+C content | 0.56796 |
| Mean single sequence MFE | -37.09 |
| Consensus MFE | -30.20 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18329169 115 - 22422827 CAGUCGCAGUCGCAACGGCGACGGACUUCGAUUGUUGGGGUAUUAUUAGACAUCCAUGGGAAUGGACAUCCUCUGCUCGAGUCCACCACCACUUGCACCGCCGAGUGGUGCAUCA ..(((((.((....)).)))))(((((.(((..((.((((............(((((....)))))...)))).))))))))))..(((((((((......)))))))))..... ( -40.56, z-score = -1.32, R) >droSec1.super_8 632943 95 - 3762037 CAGUCGCAGCCGCGACCACGACGGGCUUCGAUUGUGGGCGUAUUAUUAGACAUCCAUAGGAAUGGACAUCC--------------------CGUGCACCACCGAGUGGUGCAUCA ((((((.((((.((.......)))))).)))))).(((.((........)).(((((....)))))...))--------------------)(((((((((...))))))))).. ( -34.80, z-score = -1.71, R) >droSim1.chrX 14142545 95 - 17042790 CAGUCGCAGCCGCGACCACGACGGGCUUCGAUUGUGGGCGUAUUAUUAGACAUCCAUGGGAAUGGACAUCC--------------------CGUGCACCACCGAGUGGUGCAUCA ((((((.((((.((.......)))))).))))))(((..((........))..))).((((.......)))--------------------)(((((((((...))))))))).. ( -35.90, z-score = -1.46, R) >consensus CAGUCGCAGCCGCGACCACGACGGGCUUCGAUUGUGGGCGUAUUAUUAGACAUCCAUGGGAAUGGACAUCC____________________CGUGCACCACCGAGUGGUGCAUCA ((((((.((((.((.......)))))).))))))..............((..(((((....)))))..))......................(((((((((...))))))))).. (-30.20 = -30.20 + 0.00)

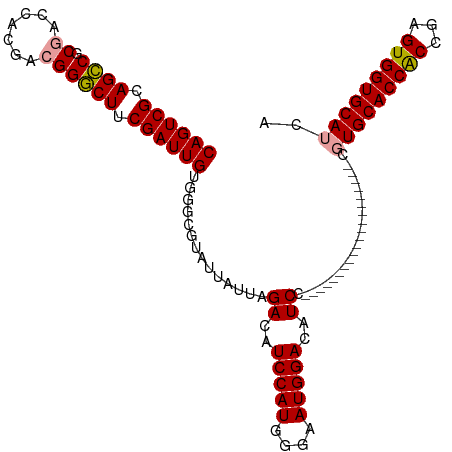
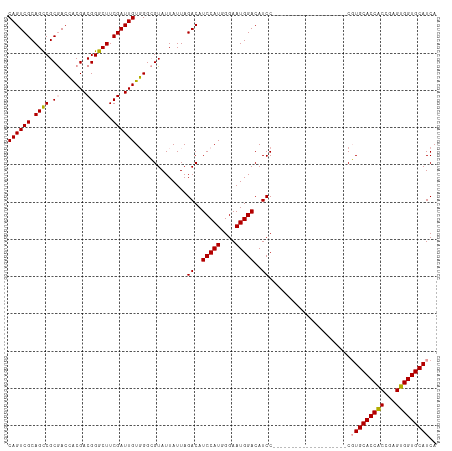
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:40 2011