| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,802,381 – 11,802,436 |
| Length | 55 |
| Max. P | 0.609313 |

| Location | 11,802,381 – 11,802,436 |
|---|---|
| Length | 55 |
| Sequences | 14 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.58894 |
| G+C content | 0.55070 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -7.93 |
| Energy contribution | -8.06 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.86 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11802381 55 + 23011544 --CGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGC------------- --.((((((((....)))))))).((.(((((((.((....)).)))).)))..)).------------- ( -22.10, z-score = -1.77, R) >droSim1.chr2L 11612632 55 + 22036055 --CGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGC------------- --.((((((((....)))))))).((.(((((((.((....)).)))).)))..)).------------- ( -22.10, z-score = -1.77, R) >droSec1.super_3 7191472 55 + 7220098 --CGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGC------------- --.((((((((....)))))))).((.(((((((.((....)).)))).)))..)).------------- ( -22.10, z-score = -1.77, R) >droYak2.chr2L 8223018 55 + 22324452 --CGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGC------------- --.((((((((....)))))))).((.(((((((.((....)).)))).)))..)).------------- ( -22.10, z-score = -1.77, R) >droAna3.scaffold_12943 2571286 55 - 5039921 --CGCACAUGGUUGACCAUGUGCUCCUUGCGCGUAAACGUCUUGGCGCAGUACUGGC------------- --.((((((((....))))))))...((((((.(((.....))))))))).......------------- ( -21.30, z-score = -1.81, R) >dp4.chr4_group3 968228 55 + 11692001 --CGCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGC------------- --.(((.((((....)))).))).((.(((((((.((....)).)))).)))..)).------------- ( -16.10, z-score = -0.33, R) >droPer1.super_5 4565123 55 + 6813705 --CGCAAGUGCACCCGCACUUGACCUGGAGUCCGAAAGCGCUUUUCGCAAUAUUGGC------------- --..(((((((....)))))))....(((((.(....).))))).............------------- ( -14.70, z-score = -0.53, R) >droEre2.scaffold_4690 2332603 67 + 18748788 --CGCAGAUGGACCUGCAUGUGCGUCUGCAGCGAGGAC-UGUGUGUGCAGCUUCUUCUUGCAGAUCUUGC --((((.(((......))).))))(((((((.((((((-(((....)))).))))).)))))))...... ( -25.80, z-score = -0.98, R) >droWil1.scaffold_180708 2438655 55 + 12563649 --CGCACAUGGUUAACCAUAUGCUCCUUGCGCGUAAACGUCUUUGCGCAGUACUGGC------------- --.(((.((((....)))).))).((.((((((((((....))))))).)))..)).------------- ( -17.60, z-score = -1.69, R) >droVir3.scaffold_12963 6814254 55 - 20206255 --CGCACAUGAUUAACCAUGUGCUCCUUGCGCGUAAACGUCUUGGCGCAGUACUGAC------------- --.(((((((......)))))))...((((((.(((.....))))))))).......------------- ( -17.80, z-score = -2.10, R) >droGri2.scaffold_15126 6465403 55 + 8399593 --CGCACAUGAUUGACCAUGUGCUCCUUGCGCGUAAACGUCUUGGCGCAGUACUGGC------------- --.(((((((......)))))))...((((((.(((.....))))))))).......------------- ( -17.80, z-score = -1.07, R) >anoGam1.chr3R 30570368 52 - 53272125 CGCACAGAUGAUACGCAGCUUGCGCGA-GCUGGAGGACAUCAUUGCGCAGGCG----------------- .((...........)).((((((((((-..((.....))...)))))))))).----------------- ( -18.60, z-score = -1.57, R) >apiMel3.GroupUn 48410928 55 + 399230636 --CGAACAUGAUUUACCAUAUGUUCUUUUCUAGUAAAUGUUUUUGAACAAUAUUGGC------------- --.(((((((........)))))))....((((((..((((....)))).)))))).------------- ( -8.50, z-score = -1.21, R) >triCas2.ChLG8 1912539 55 - 15773733 --CGCACGUGAUUCACCAUGUGUUCCUUGCGGGUGAAGGUUUUGGCACAAUACUGAC------------- --.((......((((((.((((.....)))))))))).......))...........------------- ( -11.92, z-score = 0.51, R) >consensus __CGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGC_____________ ...(((((((......)))))))............................................... ( -7.93 = -8.06 + 0.14)


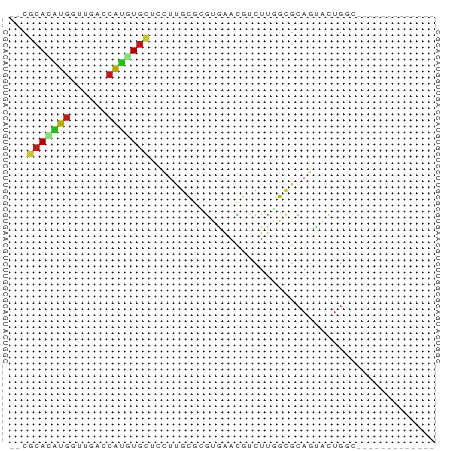
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:42 2011