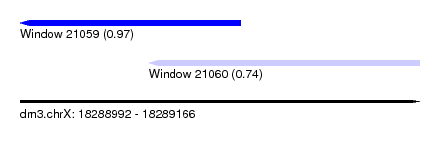
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,288,992 – 18,289,166 |
| Length | 174 |
| Max. P | 0.969410 |

| Location | 18,288,992 – 18,289,088 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Shannon entropy | 0.28697 |
| G+C content | 0.46528 |
| Mean single sequence MFE | -20.19 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.26 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
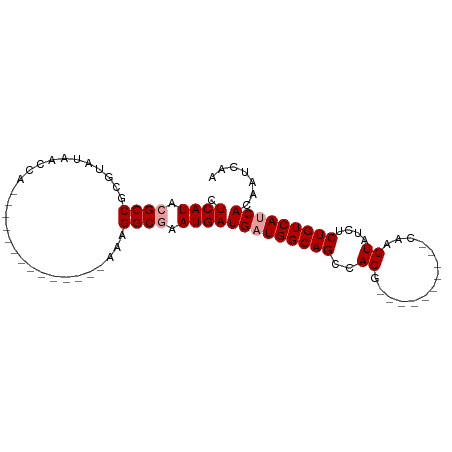
>dm3.chrX 18288992 96 - 22422827 CUCAUACGCCGCGUAUAACCAUUAAAAGGCGAACCACAGGCAAAUGAUGAUGGCAGCCACGAAAUUACAUACGAGUAUCUCUGUCAACACAAUCAA ......((((...((.......))...)))).............((((..((((((....((...(((......))))).)))))).....)))). ( -13.70, z-score = 0.78, R) >droSec1.super_8 598275 72 - 3762037 CUCAUACGCCGCGUAUACCCA--------------AAAGGCGAAUGAUGAUGGCAGCCACG----------CAAGUUUCUCUGUCAUCACAAUCAA .((((.((((...........--------------...)))).))))(((((((((..((.----------...))....)))))))))....... ( -23.44, z-score = -3.79, R) >droSim1.chrX 14102995 72 - 17042790 CUCAUACGCCGCGUAUAACCA--------------AAAGGCGAAUGAUGAUGGCAGCCACG----------CAAGUAUCUCUGUCAUCACAAUCAA .((((.((((...........--------------...)))).))))(((((((((..((.----------...))....)))))))))....... ( -23.44, z-score = -3.92, R) >consensus CUCAUACGCCGCGUAUAACCA______________AAAGGCGAAUGAUGAUGGCAGCCACG__________CAAGUAUCUCUGUCAUCACAAUCAA .((((.((((............................)))).))))(((((((((..((..............))....)))))))))....... (-15.60 = -16.26 + 0.67)

| Location | 18,289,048 – 18,289,166 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.55 |
| Shannon entropy | 0.49385 |
| G+C content | 0.46802 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -13.04 |
| Energy contribution | -11.97 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
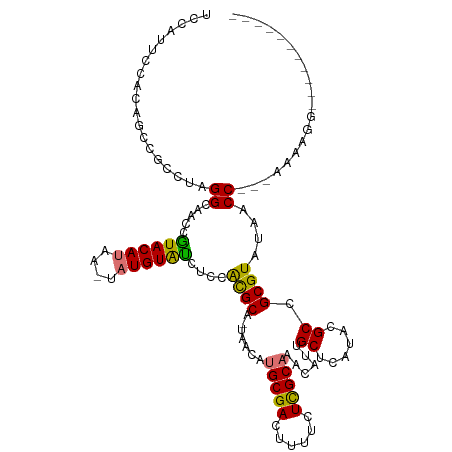
>dm3.chrX 18289048 118 - 22422827 UCCAUUCCAUAACCGCCUAGGCAACCGUACAUAA-UAUGUAUGUCUGUGCA-GAACAUGCGACUUUUCUCGCAAACAUUGCUCAUACGCCGCGUAUAACCAUUAAAAGGCGAACCACAGG .............(((((.(((....((((((..-.))))))((.((.(((-(....(((((......)))))....)))).)).)))))(.(.....))......)))))......... ( -26.80, z-score = -1.45, R) >droYak2.chrX 16917163 91 - 21770863 -------------UCCAUAGGAAAGCAUACA----UAUGUAU----AUGCA-AAACUUGCGAGUUUUCUUGCAAAUAUCGCGCAUACGCCGCGUAUAACC---AGAGUACAAACAU---- -------------......((...((((...----.))))((----((((.-....(((((((....))))))).....(((....))).))))))..))---.............---- ( -19.60, z-score = -1.33, R) >droSec1.super_8 598321 106 - 3762037 UCCAUUCCACAGCCGCCUAGGCAACCGUACAUAAGUAUGUGUCGACACGCACUCGUAUGCGACUUUUCUCGCAAACAUUGCUCAUACGCCGCGUAUACCC---AAAAGG----------- .((..((.((((((.....)))...(((((....)))))))).)).((((....((.(((((......))))).))...((......)).))))......---....))----------- ( -24.40, z-score = -1.35, R) >droSim1.chrX 14103041 106 - 17042790 UCCAUUCCACAGCCGCCUAGGCAACCGUACAUAAGUAUGUGCCUCCACGCACUCGUAUGCGACUUUUCUCGCUAACAUUGCUCAUACGCCGCGUAUAACC---AAAAGG----------- ...........(((.....)))..((((((((....))))))....((((...((((((.((......))((.......)).))))))..))))......---....))----------- ( -22.60, z-score = -1.13, R) >consensus UCCAUUCCACAGCCGCCUAGGCAACCGUACAUAA_UAUGUAUCUCCACGCA_UAACAUGCGACUUUUCUCGCAAACAUUGCUCAUACGCCGCGUAUAACC___AAAAGG___________ ...............(((........((((((....))))))....((((.......(((((......)))))......((......)).))))............)))........... (-13.04 = -11.97 + -1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:28 2011