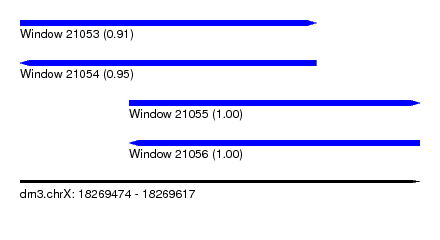
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,269,474 – 18,269,617 |
| Length | 143 |
| Max. P | 0.999060 |

| Location | 18,269,474 – 18,269,580 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.35983 |
| G+C content | 0.46381 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -20.72 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18269474 106 + 22422827 AAAUCGAACUGGAAUCGAGGG-UCUAUCAAAUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUAGGCGAUGAAUCAGCG-------------UCCAUGGAACUGCUUCAUCGCCUAACUGCA ...((((....((.((((...-.((.((.....))))...)))).)).))))((......(((((((((((.(((..-------------((....)).))).)))))))))))...)). ( -31.50, z-score = -1.71, R) >droEre2.scaffold_4690 8590196 119 + 18748788 AAAUCGAACUGGAAUCGAGGGGUCUUUCAACUUUAAGUUCUCAAGUUCUCGAGCUGGAUUUAGGCGAUGAAUCUGCGGC-ACGUGGCGCAUUCAUGGGACUUAUUCAUCGUCUAUCUACA .....((((((((((..((((.........))))..)))))..)))))....(.((((..((((((((((((.((((.(-....).)))).((....))...))))))))))))))))). ( -31.80, z-score = -0.90, R) >droYak2.chrX 16897467 113 + 21770863 AAAUCGAACUAGAAUCGAGGGGUCCUUCAACUUGAAGUUCCCAUGCUG-------GGAUUUAGACGAUGAAUCAGCGGCCACUUGGAGCAUUAGUGGAACUGCUUCAUCGUCUAGCUUCA ...((((.......)))).(((..((((.....))))..)))......-------(((.((((((((((((.(((...(((((.........)))))..))).)))))))))))).))). ( -41.50, z-score = -3.32, R) >droSec1.super_8 578967 106 + 3762037 AAAUCGAACUGGAAUCGAGGG-UCUAACAACUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUGGGCGAUGAAUCAGCG-------------UCCAUGGAACUGCUUCAUCGCCUACCUGCA ...((((..((((........-))))..(((((((.....))))))).))))((.....((((((((((((.(((..-------------((....)).))).))))))))))))..)). ( -35.90, z-score = -2.74, R) >droSim1.chrX 14087622 106 + 17042790 AAAUCGAACUGGAAUCGGGGG-UCUAUCAACUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUGGGCGAUGAAUCAGCG-------------UCCAUGGAACUGCUUCAUCGCCUACCUGCA ...((((.(((....)))...-......(((((((.....))))))).))))((.....((((((((((((.(((..-------------((....)).))).))))))))))))..)). ( -36.20, z-score = -2.40, R) >consensus AAAUCGAACUGGAAUCGAGGG_UCUAUCAACUUGAAGUUAUCGAGUUAUCGAGCUGAAUGUAGGCGAUGAAUCAGCG_____________UCCAUGGAACUGCUUCAUCGCCUACCUGCA ...((((.......))))..........(((((((.....)))))))............((((((((((((.(((........................))).))))))))))))..... (-20.72 = -21.32 + 0.60)

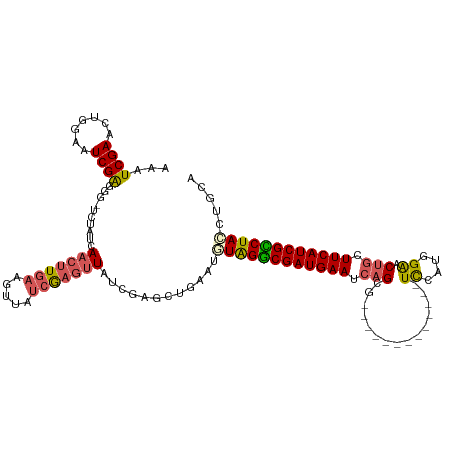
| Location | 18,269,474 – 18,269,580 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.35983 |
| G+C content | 0.46381 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -18.15 |
| Energy contribution | -19.31 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18269474 106 - 22422827 UGCAGUUAGGCGAUGAAGCAGUUCCAUGGA-------------CGCUGAUUCAUCGCCUACAUUCAGCUCGAUAACUCGAUAACUUCAAUUUGAUAGA-CCCUCGAUUCCAGUUCGAUUU ((.((((((((((((((.((((((....))-------------.)))).)))))))))..........((((....))))))))).))..........-...((((.......))))... ( -30.00, z-score = -2.46, R) >droEre2.scaffold_4690 8590196 119 - 18748788 UGUAGAUAGACGAUGAAUAAGUCCCAUGAAUGCGCCACGU-GCCGCAGAUUCAUCGCCUAAAUCCAGCUCGAGAACUUGAGAACUUAAAGUUGAAAGACCCCUCGAUUCCAGUUCGAUUU ......(((.((((((((..((..((((.........)))-)..))..)))))))).)))((((....((((....))))(((((...((((((........))))))..))))))))). ( -24.90, z-score = -1.24, R) >droYak2.chrX 16897467 113 - 21770863 UGAAGCUAGACGAUGAAGCAGUUCCACUAAUGCUCCAAGUGGCCGCUGAUUCAUCGUCUAAAUCC-------CAGCAUGGGAACUUCAAGUUGAAGGACCCCUCGAUUCUAGUUCGAUUU (((((.(((((((((((.((((.(((((.........)))))..)))).)))))))))))..(((-------(.....)))).)))))((((((.(....).))))))............ ( -39.20, z-score = -3.73, R) >droSec1.super_8 578967 106 - 3762037 UGCAGGUAGGCGAUGAAGCAGUUCCAUGGA-------------CGCUGAUUCAUCGCCCACAUUCAGCUCGAUAACUCGAUAACUUCAAGUUGUUAGA-CCCUCGAUUCCAGUUCGAUUU ....(((.(((((((((.((((((....))-------------.)))).))))))))).........((.(((((((.((.....)).))))))))))-)).((((.......))))... ( -32.50, z-score = -2.66, R) >droSim1.chrX 14087622 106 - 17042790 UGCAGGUAGGCGAUGAAGCAGUUCCAUGGA-------------CGCUGAUUCAUCGCCCACAUUCAGCUCGAUAACUCGAUAACUUCAAGUUGAUAGA-CCCCCGAUUCCAGUUCGAUUU ....(((.(((((((((.((((((....))-------------.)))).))))))))).....((((((.((.............)).))))))...)-))..(((.......))).... ( -30.62, z-score = -2.30, R) >consensus UGCAGGUAGGCGAUGAAGCAGUUCCAUGGA_____________CGCUGAUUCAUCGCCUACAUUCAGCUCGAUAACUCGAUAACUUCAAGUUGAUAGA_CCCUCGAUUCCAGUUCGAUUU ......(((((((((((.((((......................)))).)))))))))))................((((..(((...((((((........))))))..)))))))... (-18.15 = -19.31 + 1.16)

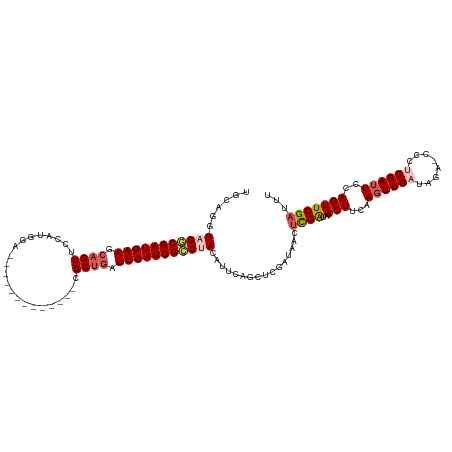
| Location | 18,269,513 – 18,269,617 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Shannon entropy | 0.31586 |
| G+C content | 0.44533 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -28.89 |
| Energy contribution | -28.82 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18269513 104 + 22422827 UCGAGUUAUCGAGCUGAAUGUAGGCGAUGAAUCAGCG------------UCCAUGGAACUGCUUCAUCGCCUAACUGCAGCAUAUGGACUCAAAUGAUUGUAAUUGAUAACUUUGA ..((((((((((((((....(((((((((((.(((..------------((....)).))).)))))))))))....)))).((..(.(......).)..)).))))))))))... ( -40.30, z-score = -5.18, R) >droEre2.scaffold_4690 8590236 108 + 18748788 UCAAGUUCUCGAGCUGGAUUUAGGCGAUGAAUCUGCGGCACGUGGCGCAUUCAUGGGACUUAUUCAUCGUCUAUCUACAGCAUGCAG---CAUAUA-----AAUUGAUCAUUUUGA (((((...((((((((....((((((((((((.((((.(....).)))).((....))...))))))))))))....))))(((...---)))...-----..))))....))))) ( -29.30, z-score = -0.83, R) >droSec1.super_8 579006 104 + 3762037 UCGAGUUAUCGAGCUGAAUGUGGGCGAUGAAUCAGCG------------UCCAUGGAACUGCUUCAUCGCCUACCUGCAGCAUAUGAACUCAAAUGCUUGUAAUUGAUCACUUUGA ..((((.(((((.......((((((((((((.(((..------------((....)).))).)))))))))))).((((((((.((....)).)))).)))).))))).))))... ( -38.30, z-score = -4.02, R) >droSim1.chrX 14087661 104 + 17042790 UCGAGUUAUCGAGCUGAAUGUGGGCGAUGAAUCAGCG------------UCCAUGGAACUGCUUCAUCGCCUACCUGCAGCAUAUGAACUCAAAUGCUUGUAAUCGAUCACUUCGA ((((((.(((((.......((((((((((((.(((..------------((....)).))).)))))))))))).((((((((.((....)).)))).)))).))))).)).)))) ( -42.10, z-score = -4.88, R) >consensus UCGAGUUAUCGAGCUGAAUGUAGGCGAUGAAUCAGCG____________UCCAUGGAACUGCUUCAUCGCCUACCUGCAGCAUAUGAACUCAAAUGCUUGUAAUUGAUCACUUUGA ((((((.(((((((((...((((((((((((.(((.......................))).))))))))))))...)))).(((........))).......))))).)).)))) (-28.89 = -28.82 + -0.06)

| Location | 18,269,513 – 18,269,617 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Shannon entropy | 0.31586 |
| G+C content | 0.44533 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -23.59 |
| Energy contribution | -26.40 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18269513 104 - 22422827 UCAAAGUUAUCAAUUACAAUCAUUUGAGUCCAUAUGCUGCAGUUAGGCGAUGAAGCAGUUCCAUGGA------------CGCUGAUUCAUCGCCUACAUUCAGCUCGAUAACUCGA ....(((((((.....(((....))).........((((.((((((((((((((.((((((....))------------.)))).))))))))))).)))))))..)))))))... ( -37.40, z-score = -5.14, R) >droEre2.scaffold_4690 8590236 108 - 18748788 UCAAAAUGAUCAAUU-----UAUAUG---CUGCAUGCUGUAGAUAGACGAUGAAUAAGUCCCAUGAAUGCGCCACGUGCCGCAGAUUCAUCGCCUAAAUCCAGCUCGAGAACUUGA ...............-----......---......((((.(..(((.((((((((..((..((((.........))))..))..)))))))).)))..).))))((((....)))) ( -21.10, z-score = 0.24, R) >droSec1.super_8 579006 104 - 3762037 UCAAAGUGAUCAAUUACAAGCAUUUGAGUUCAUAUGCUGCAGGUAGGCGAUGAAGCAGUUCCAUGGA------------CGCUGAUUCAUCGCCCACAUUCAGCUCGAUAACUCGA .....((((....))))......(((((((.....((((...((.(((((((((.((((((....))------------.)))).))))))))).))...)))).....))))))) ( -36.50, z-score = -3.67, R) >droSim1.chrX 14087661 104 - 17042790 UCGAAGUGAUCGAUUACAAGCAUUUGAGUUCAUAUGCUGCAGGUAGGCGAUGAAGCAGUUCCAUGGA------------CGCUGAUUCAUCGCCCACAUUCAGCUCGAUAACUCGA ((((.((.(((((.....(((((.((....)).)))))((..((.(((((((((.((((((....))------------.)))).))))))))).)).....))))))).)))))) ( -38.90, z-score = -3.71, R) >consensus UCAAAGUGAUCAAUUACAAGCAUUUGAGUCCAUAUGCUGCAGGUAGGCGAUGAAGCAGUUCCAUGGA____________CGCUGAUUCAUCGCCCACAUUCAGCUCGAUAACUCGA ........................((((((.....((((...((((((((((((.((((.....................)))).))))))))))))...)))).....)))))). (-23.59 = -26.40 + 2.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:25 2011