| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,239,319 – 18,239,386 |
| Length | 67 |
| Max. P | 0.977960 |

| Location | 18,239,319 – 18,239,386 |
|---|---|
| Length | 67 |
| Sequences | 10 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Shannon entropy | 0.52045 |
| G+C content | 0.43189 |
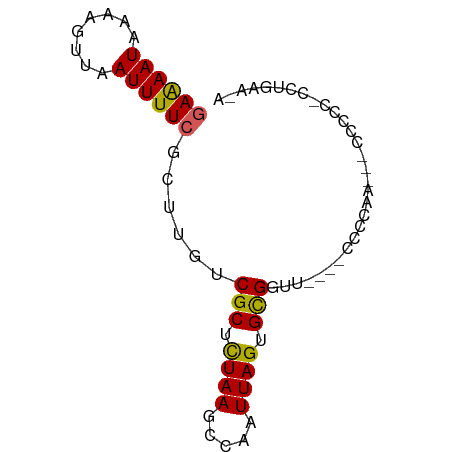
| Mean single sequence MFE | -12.18 |
| Consensus MFE | -8.22 |
| Energy contribution | -7.84 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.977960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
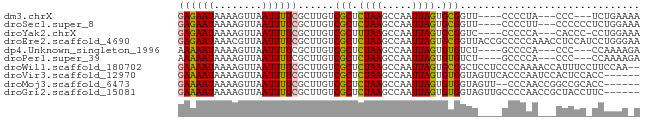
>dm3.chrX 18239319 67 + 22422827 GAGAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGCGGUU----CCCCUA---CCC---UCUGAAAA ((((((........))))))(((((......)))))...((((.(.(((.----.....)---)))---.))))... ( -12.00, z-score = -1.48, R) >droSec1.super_8 551283 70 + 3762037 GAGAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGCGGUU----CCCCUU---CCCCCCUCUGGAAA ((((((........)))))).....((((.((((.....)))).))))..----....((---((.......)))). ( -12.50, z-score = -0.92, R) >droYak2.chrX 16873009 69 + 21770863 GAGAAUAAAAGUUAAUUUUCGCUUGUCGCUUUAAGCCAAUUAGUGCGGUC----CCCCCA---CACCC-CCUGGAAA ((((((........))))))(((((......)))))......(((.((..----..))))---).((.-...))... ( -11.60, z-score = -0.68, R) >droEre2.scaffold_4690 8562494 77 + 18748788 GAGAAUAAACGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGCGGUUACCGCCCCCAAACCUCCAUCCUGGGAA ((((((........))))))((...((((.((((.....)))).)))).....)).((((...........)))).. ( -14.80, z-score = -0.49, R) >dp4.Unknown_singleton_1996 266 67 + 554 AAAAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGUGUCU----GCCCCA---CCC---CCAAAAGA ....................((..(.(((.((((.....)))).))).).----))....---...---........ ( -8.00, z-score = -1.59, R) >droPer1.super_39 214726 67 - 745454 AAAAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGUGUCU----GCCCCA---CCC---CCAAAAGA ....................((..(.(((.((((.....)))).))).).----))....---...---........ ( -8.00, z-score = -1.59, R) >droWil1.scaffold_180702 1480326 75 + 4511350 GAAAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGCGGCUCCUCCCCAAAACCAUUUCCUUCCAA-- ((((((........))))))....(((((.((((.....)))).)))))..........................-- ( -13.90, z-score = -3.21, R) >droVir3.scaffold_12970 3011608 71 + 11907090 GAAAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGUGGUAGUUCACCCAAUCCACUCCACC------ ((((((........))))))(((((......)))))........((((.(((...........))))))).------ ( -11.00, z-score = -1.84, R) >droMoj3.scaffold_6473 3828071 69 + 16943266 GAAAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGUGGUAGUU--CCCAACCGGCCGCACC------ ((((((........))))))(((((......)))))......(((((((.((.--....))..))))))).------ ( -15.70, z-score = -1.99, R) >droGri2.scaffold_15081 3442511 71 - 4274704 GAAAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGUGGUAGUUGCCCCAACCGCUACCUUC------ ((((((........))))))(((((......))))).....((.((((..((((...))))..))))))..------ ( -14.30, z-score = -2.12, R) >consensus GAAAAUAAAAGUUAAUUUUCGCUUGUCGCUCUAAGCCAAUUAGUGCGGUU____CCCCAA___CCCCC_CCUGAA_A ((((((........))))))......(((.((((.....)))).))).............................. ( -8.22 = -7.84 + -0.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:18 2011