
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,787,672 – 11,787,776 |
| Length | 104 |
| Max. P | 0.746537 |

| Location | 11,787,672 – 11,787,776 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.61 |
| Shannon entropy | 0.65352 |
| G+C content | 0.42183 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -10.05 |
| Energy contribution | -12.59 |
| Covariance contribution | 2.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11787672 104 - 23011544 -----------GGAGCGCCUUUUCGACCC---GAAAAUUGUUCUUUCUGUGGGGCAGUUUUC-AGCUCGACUUUGUCCCCAUGGAAAGUCAUAAUACCCUGAAUAUUUUGAGAUCAUAC -----------((..((......))..))---((..(((((.(((((((((((((((...((-.....))..))).))))))))))))..)))))...((.((....)).)).)).... ( -24.90, z-score = -0.66, R) >droSim1.chr2L 11597527 104 - 22036055 -----------GGAGCGCCUUUUCGACCC---GAAAAUUGUUCUUUAUGUGGGGCAGUUUUC-AGCUCGACUUUCUCCCCAUGGAAAGUCAUAAUACCCUGAAUAUUUUCAGAUCAUAU -----------.((((..(.....)....---((((((((((((......))))))))))))-.))))((((((((......))))))))........(((((....)))))....... ( -31.30, z-score = -3.16, R) >droSec1.super_3 7176742 104 - 7220098 -----------GGAGCGCCUUUUCGACCC---GAAAAUUGUUCUUUAUGUGGUGCAGUUUUC-AGCUCGACUUUCUCCCCAUGGAAAGUCAUAAUACCCUGAAUAUUUUGAGAUCAUAU -----------.((((..(.....)....---(((((((((.((......)).)))))))))-.))))((((((((......))))))))........((.((....)).))....... ( -24.10, z-score = -1.45, R) >droYak2.chr2L 8208353 105 - 22324452 -----------GAAGCGCCUUUUCGACCC---GAAAAUUGUUCUUUAUGUGGGGCAUUUUUUAAGCUCGACUUUCUCCCCAUGGAAAGUCAUAAUACCCUAAAUAUUUUGAGACCAUAU -----------((((.((.((((((...)---)))))..)).))))(((((((((.........))).((((((((......)))))))).......................)))))) ( -20.70, z-score = -0.86, R) >droEre2.scaffold_4929 13014640 103 + 26641161 -----------GGAGCGCCUUUUCAACCC---GAAAAUUGUUCUUUAUGUGGGGCAGUUUUC-AGCUAGACUUUCUCUCCAUGGAAAGUCAUAAUACCCUGAAUAUUUUGAGAUCACA- -----------((....)).((((((...---((((((((((((......))))))))))))-.....((((((((......)))))))).................)))))).....- ( -26.10, z-score = -1.47, R) >droAna3.scaffold_12916 6689555 106 - 16180835 GCAGCUCGUUUUUUCCCCAUUUUGCACCCUCUGCAAAUUUGACUUUGUGUGGGGCAGUUUUC-UGCAUGACUUCUUC---------AGUUUAAAUACCCUGCAUGUAUCCUCAAAA--- ((((...........(((((((((((.....))))))...........)))))((((....)-)))...........---------............))))..............--- ( -20.30, z-score = -0.45, R) >droMoj3.scaffold_6500 25529371 82 - 32352404 --------------------------------GACGUUCAUUCAUUACAAAAAACAGCUCAAACGAAGGGCAGCUGU----GGGGAGCAUGUAGUAGGUCCAAU-UUUUGAGCUCCCAU --------------------------------.....................((((((....(....)..))))))----.((((((................-......)))))).. ( -18.65, z-score = -0.40, R) >consensus ___________GGAGCGCCUUUUCGACCC___GAAAAUUGUUCUUUAUGUGGGGCAGUUUUC_AGCUCGACUUUCUCCCCAUGGAAAGUCAUAAUACCCUGAAUAUUUUGAGAUCAUAU ................................((((((((((((......))))))))))))......((((((((......))))))))............................. (-10.05 = -12.59 + 2.54)

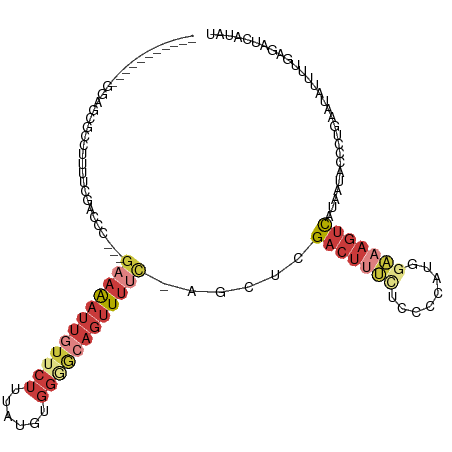
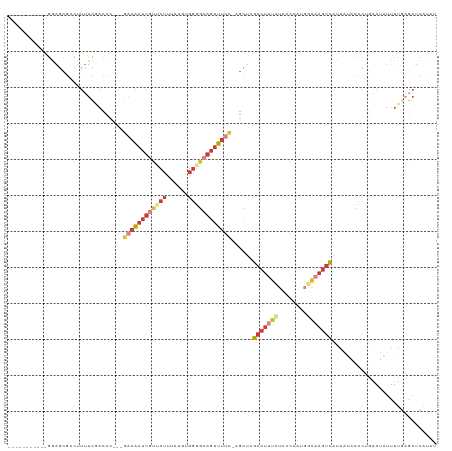
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:41 2011