
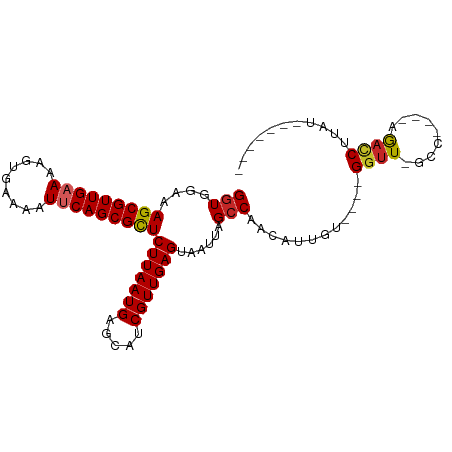
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,775,934 – 11,776,054 |
| Length | 120 |
| Max. P | 0.729192 |

| Location | 11,775,934 – 11,776,027 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.27927 |
| G+C content | 0.43309 |
| Mean single sequence MFE | -23.39 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 11775934 93 + 23011544 GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGU----GGUU-GCC----AGACCUUAUCCCUUGU (((((..(((((((((.........)))))))))(((((((.....)))))))....((((((......)----))))-)))----..)))........... ( -23.10, z-score = -1.15, R) >droSim1.chr2L 11585933 86 + 22036055 GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGU----GGUU-GCC----AGACCUUAU------- (((((..(((((((((.........)))))))))(((((((.....)))))))....((((((......)----))))-)))----..)))....------- ( -23.10, z-score = -1.77, R) >droSec1.super_3 7165237 86 + 7220098 GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGU----GGUU-GCC----AGACCUUAU------- (((((..(((((((((.........)))))))))(((((((.....)))))))....((((((......)----))))-)))----..)))....------- ( -23.10, z-score = -1.77, R) >droYak2.chr2L 8196267 93 + 22324452 GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUCGC----GGUU-GCC----AGACCUUAUCCCUAUU ((.(((..((((((((.........))))))((((.....))))...((((.((.....))))))...))----((((-...----.))))...)))))... ( -24.10, z-score = -1.42, R) >droEre2.scaffold_4929 13003156 93 - 26641161 GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGC----GGUU-GCC----AGACCUUAUCCCUAUG ((.(((.(((((((((.........)))))))))........(((..((((.((.....))))))..)))----((((-...----.))))...)))))... ( -25.00, z-score = -1.76, R) >droAna3.scaffold_12916 9483388 86 - 16180835 GUUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGU----GGUUCGCC----AGACCUUA-------- (((((..(((((((((.........)))))))))(((((((.....))))))).......)))))..(.(----((....))----).).....-------- ( -25.00, z-score = -2.44, R) >dp4.chr4_group3 2648098 85 + 11692001 GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGU----GGUU-GCA----AGACCUUC-------- (((....(((((((((.........)))))))))(((((((.....)))))))....((((((......)----))))-)..----..)))...-------- ( -22.40, z-score = -1.58, R) >droPer1.super_1 4122578 85 + 10282868 GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGU----GGUU-GCA----AGACCUUC-------- (((....(((((((((.........)))))))))(((((((.....)))))))....((((((......)----))))-)..----..)))...-------- ( -22.40, z-score = -1.58, R) >droVir3.scaffold_12963 6370913 94 + 20206255 GGUGCAAAGCGUUGAAAAGUGAAAAUGCAGCGUUCUUAAUGAGCAGCGUUGAGUAAUUAGCCAACAUUGUGUGCGGUUUCCU----CAAGCUCUUCAU---- .......(((((((....((....)).)))))))....(((((.(((.(((((.....((((.((.....))..))))..))----)))))).)))))---- ( -22.80, z-score = 0.02, R) >droMoj3.scaffold_6500 25513751 100 + 32352404 GGUGCAAAGCGUUGAAAAGUGAAAAUGCAGCGUUCUUAAUGAGCACCGUUGAGUAAUUAGCCAACAUUGUUCGCGGCUUCCUUCCCCAAUCUCGCCUCUC-- (((((...((((((....((....)).))))))((.....)))))))((((.((.....)))))).........(((................)))....-- ( -22.89, z-score = -0.56, R) >consensus GGUGGAAAGCGUUGAAAAGUGAAAAUUCAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGU____GGUU_GCC____AGACCUUAU_______ (((....(((((((((.........)))))))))(((((((.....)))))))......)))............((((.........))))........... (-17.52 = -17.62 + 0.10)

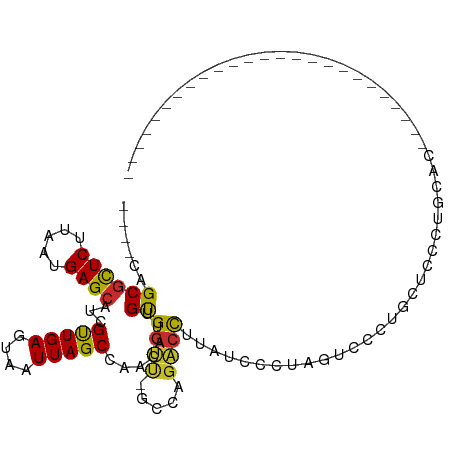
| Location | 11,775,961 – 11,776,054 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 68.35 |
| Shannon entropy | 0.57340 |
| G+C content | 0.47946 |
| Mean single sequence MFE | -17.72 |
| Consensus MFE | -10.45 |
| Energy contribution | -10.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11775961 93 + 23011544 ----CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGUGGUU-GCCAGACCUUAUCCCUUGUCCCUUGUCCCUUAUCCCUAUUCCCUAU------------- ----..((((((.....))))...((((.((.....))))))...))((((-....))))......................................------------- ( -13.70, z-score = -0.52, R) >droSim1.chr2L 11585960 86 + 22036055 ----CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGUGGUU-GCCAGACCUUAUCCCUUGUCCCUUAUCCCUAUUCCCUAU-------------------- ----..((((((.....))))...((((.((.....))))))...))((((-....))))...............................-------------------- ( -13.70, z-score = -0.71, R) >droSec1.super_3 7165264 86 + 7220098 ----CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGUGGUU-GCCAGACCUUAUCCCUUGUCCCUUAUCCCUAUUCCCUAU-------------------- ----..((((((.....))))...((((.((.....))))))...))((((-....))))...............................-------------------- ( -13.70, z-score = -0.71, R) >droEre2.scaffold_4929 13003183 106 - 26641161 ----CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGCGGUU-GCCAGACCUUAUCCCUAUGCCCUAUUCCCUGCCCCCUGCCCCUAUCCCGUAUCCCGUAU ----..((((((.....))))...((((.((.....))))))...))((((-....))))................................................... ( -16.30, z-score = -0.18, R) >droAna3.scaffold_12916 9483415 74 - 16180835 ----CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGUGGUUCGCCAGACCUUAUCCUUGUACCGACCG--------------------------------- ----....((((.....))))...((((.(((...((.......(.(((....))).).......))..)))))))..--------------------------------- ( -14.34, z-score = -0.47, R) >dp4.chr4_group3 2648125 80 + 11692001 ----CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGUGGUU-GCAAGACCUUCAUCGUAUACGAGGGCUCCUGCA-------------------------- ----((((((((.....))))...)))).......(((((......)))))-(((.(((((((.........))))).)).))).-------------------------- ( -21.60, z-score = -0.78, R) >droPer1.super_1 4122605 80 + 10282868 ----CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGUGGUU-GCAAGACCUUCAUCGUAUACGGGGGCUCCUGCA-------------------------- ----((((((((.....))))...)))).......(((((......)))))-(((.(((((((.........))))).)).))).-------------------------- ( -20.60, z-score = -0.10, R) >droWil1.scaffold_180772 3655452 80 + 8906247 CAGCGAGCUCUCUUAAUGAGCAUCGCUGAGUAAUUAGCCAACAUUGUGGUC--UUCGUCUUGAUUCU--GGCCAUGCUCCUGGU--------------------------- ((((((((((.......)))).))))))........((((.....((((((--.((.....))....--)))))).....))))--------------------------- ( -25.70, z-score = -1.96, R) >droVir3.scaffold_12963 6370940 79 + 20206255 ----CAGCGUUCUUAAUGAGCAGCGUUGAGUAAUUAGCCAACAUUGUGUGC--GGUUUCCUCAAGCU--CUUCAUCCUCUCCGCACA------------------------ ----..(((......(((((.(((.(((((.....((((.((.....))..--))))..))))))))--.)))))......)))...------------------------ ( -19.80, z-score = -1.64, R) >consensus ____CAGCGCUCUUAAUGAGCAUCGUUGAGUAAUUAGCCAACAUUGUGGUU_GCCAGACCUUAUCCCUAGUCCCUGCUCCCUGCAC_________________________ ......((((((.....))))...((((.((.....))))))...))((((.....))))................................................... (-10.45 = -10.46 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:40 2011