| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,201,148 – 18,201,241 |
| Length | 93 |
| Max. P | 0.739195 |

| Location | 18,201,148 – 18,201,241 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.35 |
| Shannon entropy | 0.51743 |
| G+C content | 0.49269 |
| Mean single sequence MFE | -19.81 |
| Consensus MFE | -11.60 |
| Energy contribution | -10.45 |
| Covariance contribution | -1.15 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
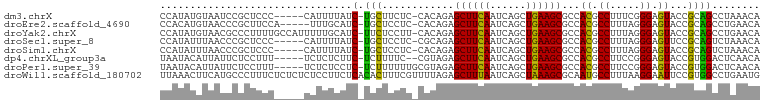
>dm3.chrX 18201148 93 - 22422827 CCAUAUGUAAUCCGCUCCC-----CAUUUUAUC-UGCUUCUC-CACAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUUCGGGAGUACCGCAGCCUAAACA .............((((((-----.......((-((......-..))))((((((......))))))............))))))............... ( -21.20, z-score = -1.62, R) >droEre2.scaffold_4690 8527343 93 - 18748788 CCACAUGUAACCCGCUUCCA-----UUUGCAUC-UGCUCCUC-CACAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUUAGGGAGUACCGCAGCCUGAACA .((..(((.....((((((.-----...((.((-((......-..))))((((((......)))))).....)).....))))))...)))...)).... ( -20.00, z-score = 0.05, R) >droYak2.chrX 16835113 98 - 21770863 CCAUAUGUAACGCCCUUUUGCCAUUUUUGCAUC-UUCUCCUU-CACAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUUAGGGAGUACCGCAGCCUGAACA .((..(((.((.((((...((............-.(((....-...)))((((((......)))))).....))....)))).))...)))...)).... ( -16.70, z-score = 1.06, R) >droSec1.super_8 513236 93 - 3762037 CCAUAUUUAACCCGCUCCC-----CAUUUUAUC-UGCUCCUC-CGCAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUUAGGGAGUUCCGCAGUCUAAACA .............((((((-----.......((-(((.....-.)))))((((((......))))))............))))))............... ( -24.40, z-score = -2.92, R) >droSim1.chrX 14022585 93 - 17042790 CCAUAUUUAACCCGCUCCC-----CAUUUUAUC-UGCUCCUC-CACAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUUAGGGAGUACCGCAGUCUAAACA .............((((((-----.......((-((......-..))))((((((......))))))............))))))............... ( -21.10, z-score = -2.29, R) >dp4.chrXL_group3a 165882 92 - 2690836 UAAUACAUUAUUCUCCUUU-----UCUCUCUUC-UCUUUUC--CGUAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUCCGGGAGUACCGUGGACUCAACA ...................-----.........-.......--.((.((((((((......)))))).(((((.(((....)))...)))))..)).)). ( -17.30, z-score = -0.66, R) >droPer1.super_39 176570 94 + 745454 UAAUACAUUAUUCUCCUUU-----UCUCUCCUC-UCUUUUUUGCGUAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUCCGGGAGUACCGUGGACUCAACA .............(((...-----...(((((.-........((((.(.((((((......)))))).).)))).....)))))......)))....... ( -21.84, z-score = -1.86, R) >droWil1.scaffold_180702 1419682 100 - 4511350 UUAAACUUCAUGCCCUUUCUCUCUCUCCUUCUCACACUUUCGUUUUAGAGCUUUAAUCAGCUAAAGCGCAAUGCCUUUAAGGAAUUCCGUGGCCUGAAUG ......((((.(((...........(((((..........((((((..((((......))))))))))..........))))).......))).)))).. ( -15.92, z-score = -0.36, R) >consensus CCAUAUGUAACCCGCUUCC_____CAUUUCAUC_UGCUCCUC_CACAGAGCUUCAAUCAGCUGAAGCGCCACGCCUUUAGGGAGUACCGCAGCCUAAACA .............(((.................................((((((......))))))((.((.((.....)).))...)))))....... (-11.60 = -10.45 + -1.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:06 2011