| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,195,074 – 18,195,166 |
| Length | 92 |
| Max. P | 0.990094 |

| Location | 18,195,074 – 18,195,166 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 68.52 |
| Shannon entropy | 0.65248 |
| G+C content | 0.32802 |
| Mean single sequence MFE | -13.00 |
| Consensus MFE | -8.50 |
| Energy contribution | -8.51 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
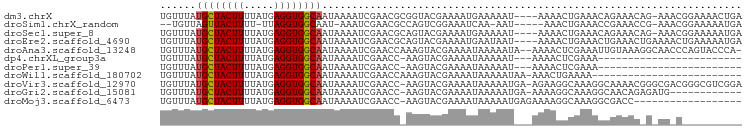
>dm3.chrX 18195074 92 - 22422827 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACGCGGUACGAAAAUGAAAAAU----AAAACUGAAACAGAAACAG-AAACGGAAAACUGA (((((.(((((((((...))))))))).....((((....))))...............----.......))))).......-...(((....))). ( -13.70, z-score = -1.47, R) >droSim1.chrX_random 4736716 86 - 5698898 --UGUUAGUUACUUUU-UUAGGUGGCAAU-AAAUCGAACGCCAGUCGGAAAUCAA-AAU-----AAACUGAAACCGAAACCG-AAACGGAAAAAUGA --.(((.(((((((..-..)))))))...-........((....((((...(((.-...-----....)))..))))...))-.))).......... ( -12.90, z-score = 0.28, R) >droSec1.super_8 507139 92 - 3762037 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACGCAGUACGAAAAUGAAAAAU----AAAACUGAAACAGAAACAG-AAACGGAAAAAUGA .((((.(((((((((...)))))))))......((.....((((...............----...)))).....)).....-)))).......... ( -13.37, z-score = -1.63, R) >droEre2.scaffold_4690 8521148 93 - 18748788 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACGCAGUACGAAAAUGAAUAAU----AAAACUGAAACUGAAACUGAAAACUGAAAAAUGA .((((.(((((((((...)))))))))......(((....((((...............----...))))....)))......)))).......... ( -12.57, z-score = -1.84, R) >droAna3.scaffold_13248 3811403 94 + 4840945 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACCAAAGUACGAAAAUAAAAAUA--AAAACUCGAAAUUGUAAAGGCAACCCAGUACCCA- .((((.(((((((((...)))))))))......(((.((....)).)))...........--.))))........(((..((....))..)))...- ( -14.10, z-score = -0.66, R) >dp4.chrXL_group3a 158583 69 - 2690836 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACC-AAGUACGAAAAUAAAAAU---AAAACUCGAAA------------------------ .((((.(((((((((...)))))))))......(((.((.-..)).)))..........---.))))......------------------------ ( -11.10, z-score = -1.94, R) >droPer1.super_39 169444 69 + 745454 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACC-AAGUACGAAAAUAAAAAU---AAAACUCGAAA------------------------ .((((.(((((((((...)))))))))......(((.((.-..)).)))..........---.))))......------------------------ ( -11.10, z-score = -1.94, R) >droWil1.scaffold_180702 3100523 71 + 4511350 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACCAAAGUACGAAAAUAAAAAUAA-AAACUGAAAA------------------------- .((((.(((((((((...)))))))))......(((.((....)).)))............-))))......------------------------- ( -12.20, z-score = -2.86, R) >droVir3.scaffold_12970 2936847 95 - 11907090 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACC-AAGUACGAAAAUAAAAAUGA-AGAAGGCAAAGGCAAAACGGGCGACGGGCGUCGGA .((((.(((((((((...)))))))))......(((.((.-..)).)))............-.....((....)).))))...((((....)))).. ( -17.00, z-score = 0.09, R) >droGri2.scaffold_15081 3390849 83 + 4274704 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACC-AAGUACGAAAAUAAAAAUGA-AAAAGGCAAAGGCAACAGAGAUG------------ (((((.(((((((((...)))))))))......(((.((.-..)).)))............-...)))))..(....).......------------ ( -13.10, z-score = -1.24, R) >droMoj3.scaffold_6473 3745390 78 - 16943266 UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACC-AAGUACGAAAAUAAAAAUGAGAAAAGGCAAAGGCGACC------------------ (((((.(((((((((...)))))))))......(((.((.-..)).)))................))))).........------------------ ( -11.90, z-score = -0.69, R) >consensus UGUUUAUGCUACUUUUAUGAGGUGGCAAUAAAAUCGAACC_AAGUACGAAAAUAAAAAU___AAAAACCGAAACCGAAAC_G_AA_C__________ ......((((((((.....))))))))...................................................................... ( -8.50 = -8.51 + 0.01)
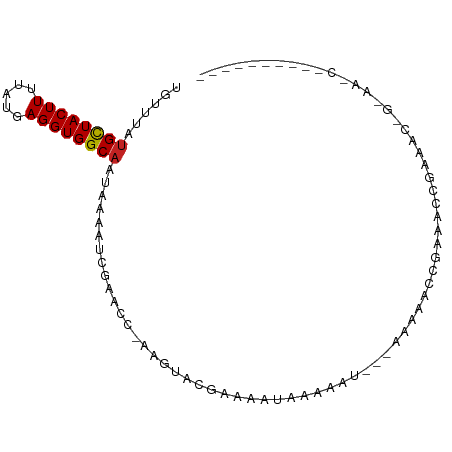
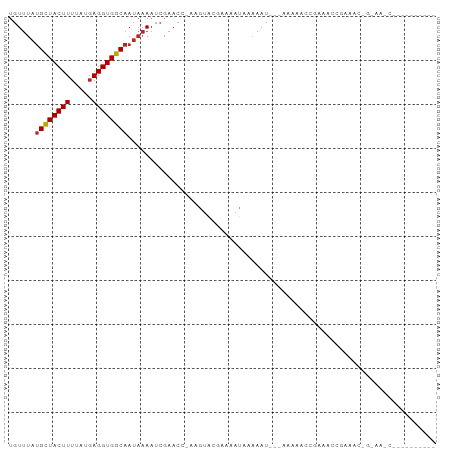
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:05 2011