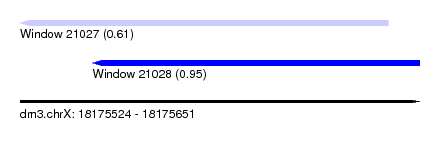
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,175,524 – 18,175,651 |
| Length | 127 |
| Max. P | 0.946111 |

| Location | 18,175,524 – 18,175,641 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.32 |
| Shannon entropy | 0.50079 |
| G+C content | 0.43248 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -15.94 |
| Energy contribution | -14.78 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
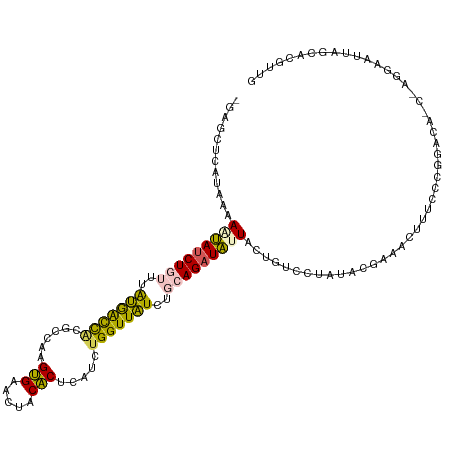
>dm3.chrX 18175524 117 - 22422827 CGAUCUCAUAAAAGUAUCUGUUUAUGGCCACGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCGGACACCAAGGAAUUAGCACGUUG .(((..(.....(((((((((..(((((((.....((((.....))))....)))))))..)))))))))..(((((................)))))....)..)))......... ( -26.29, z-score = -1.49, R) >droEre2.scaffold_4690 8502100 117 - 18748788 CGAUCUCAUAAAAGUAUCUGUUUAUGACCAUGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCGGACAUCUAGGAAUUAGCACGUUG ............(((((((((..(((((((.....((((.....))))....)))))))..)))))))))..........(((.....((((..(....)..))))......))).. ( -26.90, z-score = -1.92, R) >droYak2.chrX 16807380 117 - 21770863 UGAGCUCAUAAAAAUAGCUGUUUAUGAUCACGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCGGACACCUAGGAAUUAGCACGUUG ...(((.((...((((.((((..(((((((.....((((.....))))....)))))))..)))).))))..(((((................))))).......)).)))...... ( -20.99, z-score = 0.09, R) >dp4.chrXL_group3a 142990 104 - 2690836 -CUGUCUGUCAUAAAAUCUUAUUGGAACUGC-CAGAGCGAACUGCGCGUAUCUGGUUCUCUGCAGAUUAUUCCGUCUUGCACAGCCUCCUCC-----------AAAUUAGCACUUUG -....................(((((...((-(((((((.....)))...)))))).((.(((((((......))).)))).)).....)))-----------))............ ( -20.40, z-score = 0.02, R) >droPer1.super_39 152979 104 + 745454 -CUGUCUGUCAUAAAAUCUUAUUGGAGCUUC-CAGAGCGAACUGCGCGUAUCUGGUUCUCUGCAGAUUAUUCCGUCUUGCACAGCCUCCUCC-----------AAAUUAGCACUUUG -....................((((((...(-(((((((.....)))...)))))..((.(((((((......))).)))).))....))))-----------))............ ( -21.20, z-score = -0.15, R) >consensus _GAGCUCAUAAAAAUAUCUGUUUAUGACCACGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCGGACA_C_AGGAAUUAGCACGUUG ............(((((((((..(((((((......(((.....))).....)))))))..)))))))))............................................... (-15.94 = -14.78 + -1.16)

| Location | 18,175,547 – 18,175,651 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.49964 |
| G+C content | 0.46359 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
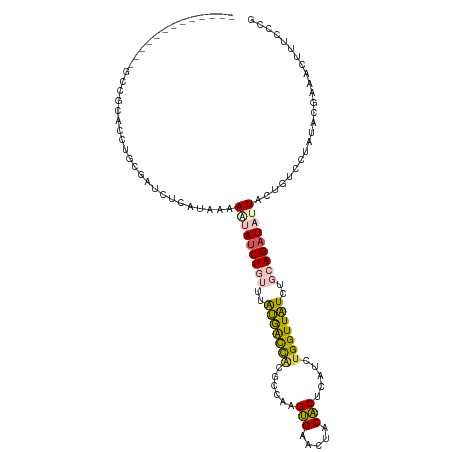
>dm3.chrX 18175547 104 - 22422827 -------------GCCGCACCUGCGAUCUCAUAAAAGUAUCUGUUUAUGGCCACGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCG -------------..(((....)))..........(((((((((..(((((((.....((((.....))))....)))))))..)))))))))........................ ( -23.90, z-score = -2.63, R) >droEre2.scaffold_4690 8502123 104 - 18748788 -------------GCCGCACCUGCGAUCUCAUAAAAGUAUCUGUUUAUGACCAUGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCG -------------..(((....)))..........(((((((((..(((((((.....((((.....))))....)))))))..)))))))))........................ ( -24.40, z-score = -3.36, R) >droYak2.chrX 16807403 104 - 21770863 -------------GCCGCACCUGUGAGCUCAUAAAAAUAGCUGUUUAUGAUCACGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCG -------------...(((...((((..(((((((........)))))))))))((((..(((.......)))..))))....))).((((....)))).................. ( -18.20, z-score = -0.59, R) >dp4.chrXL_group3a 143004 99 - 2690836 --------------CUACUCCCGCUGUCUGUCAUAAA-AUCUUAUUGGAACUGC-CAGAGCGAACUGCGCGUAUCUGGUUCUCUGCAGAUUAUUCCGUCUUGCACAGCCUCCUCC-- --------------........(((((..........-........(((...((-(((((((.....)))...))))))..)))((((((......))).)))))))).......-- ( -22.30, z-score = -1.01, R) >droPer1.super_39 152993 113 + 745454 AGUAGGCAAAGGUUCUACUCCCGCUGUCUGUCAUAAA-AUCUUAUUGGAGCUUC-CAGAGCGAACUGCGCGUAUCUGGUUCUCUGCAGAUUAUUCCGUCUUGCACAGCCUCCUCC-- (((((((....).))))))...(((((..........-........((((...(-(((((((.....)))...)))))..))))((((((......))).)))))))).......-- ( -26.60, z-score = 0.04, R) >consensus _____________GCCGCACCUGCGAUCUCAUAAAAAUAUCUGUUUAUGACCACGCCAAGUGAACUACACUCAUCUGGUUAUCUGCAGAUAUUACUGUCCUAUACGAAACUUUCCCG ...................................(((((((((..(((((((......(((.....))).....)))))))..)))))))))........................ (-14.74 = -14.46 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:56:02 2011