
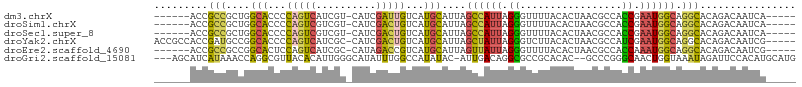
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,169,311 – 18,169,406 |
| Length | 95 |
| Max. P | 0.657245 |

| Location | 18,169,311 – 18,169,406 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.21 |
| Shannon entropy | 0.52026 |
| G+C content | 0.53281 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -14.17 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18169311 95 + 22422827 ------ACCGCCGCUGGCACCCCAGUCAUCGU-CAUCGAUUGUCAUGCAUUAGCCAUUAGGGUUUUACACUAACGCCACCGAAUGGCAGGCACAGACAAUCA----- ------..((..(((((....)))))...)).-....(((((((.(((....((((((..(((.(((...))).)))....))))))..)))..))))))).----- ( -28.60, z-score = -2.38, R) >droSim1.chrX 14011065 95 + 17042790 ------ACCGCCGCUGGCACCCCAGUCGUCGU-CAUCGACUGUCAUGCAUUAGCCAUUAGGGUUUUACACUAACGCCACCGAAUGGCAGGCACAGACAAUCA----- ------...(((....(((...((((((....-...))))))...)))....((((((..(((.(((...))).)))....)))))).)))...........----- ( -28.60, z-score = -1.81, R) >droSec1.super_8 481955 95 + 3762037 ------ACCGCCGCUGGCACCCCAGUCGUCGU-CAUCGACUGUCAUGCAUUAGCCAUUAGGGUUUUACACUAACGCCACCGAAUGGCAGGCACAGACAAUCA----- ------...(((....(((...((((((....-...))))))...)))....((((((..(((.(((...))).)))....)))))).)))...........----- ( -28.60, z-score = -1.81, R) >droYak2.chrX 16800786 101 + 21770863 ACCGCCACCGAUGCCGGCACCCCAGUCAUCGC-CAUCGACUGUCAUGCAUUAGCUAUUAGGGUCUUACACUAACGCCAUCGAAUGGCAGGCACAGACAAUCG----- ...(((...(((((.((....)).).))))((-(((((((((....((....))...)))(((.(((...))).))).))).))))).)))...........----- ( -26.20, z-score = -0.84, R) >droEre2.scaffold_4690 8496573 95 + 18748788 ------ACCGCCGCCGGCACUCCAGUCAUCGC-CAUAGACCGUCAUGCAUUAGUUAUUAGGGUUUUACACUAACGCCACCAAAUGGCAGGCACAGACAAUCG----- ------...(((...)))......(((...((-(..(((((.(.((((....).))).).))))).........((((.....)))).)))...))).....----- ( -20.00, z-score = -0.27, R) >droGri2.scaffold_15081 3363766 101 - 4274704 ---AGCAUCAUAAACCAGGCGUUACACAUUGGGCAUAUUUGGCCAUAUAC-AUUGACAGGCGCCGCACAC--GCCCGGGCAACUGGUAAAUAGAUUCCACAUGCAUG ---.((((.........((((((...((.(((.((....)).))).....-..))...))))))......--..((((....))))..............))))... ( -24.00, z-score = 0.39, R) >consensus ______ACCGCCGCCGGCACCCCAGUCAUCGU_CAUCGACUGUCAUGCAUUAGCCAUUAGGGUUUUACACUAACGCCACCGAAUGGCAGGCACAGACAAUCA_____ .........(((....(((...(((((..........)))))...)))....((((((.((.................)).)))))).)))................ (-14.17 = -13.68 + -0.49)

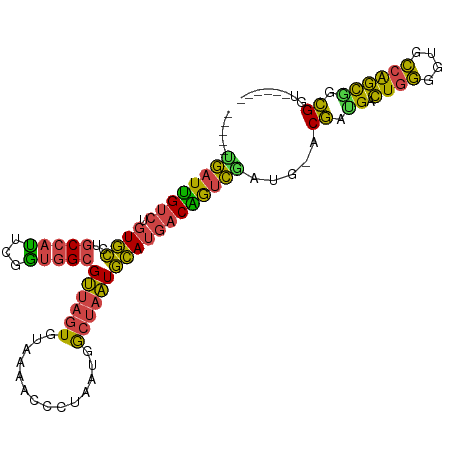
| Location | 18,169,311 – 18,169,406 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 71.21 |
| Shannon entropy | 0.52026 |
| G+C content | 0.53281 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -23.64 |
| Energy contribution | -21.56 |
| Covariance contribution | -2.08 |
| Combinations/Pair | 1.61 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.655351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18169311 95 - 22422827 -----UGAUUGUCUGUGCCUGCCAUUCGGUGGCGUUAGUGUAAAACCCUAAUGGCUAAUGCAUGACAAUCGAUG-ACGAUGACUGGGGUGCCAGCGGCGGU------ -----.(((((((.((((..((((((.(((.(((....)))...)))..))))))....)))))))))))....-.((.((.((((....)))))).))..------ ( -32.50, z-score = -1.18, R) >droSim1.chrX 14011065 95 - 17042790 -----UGAUUGUCUGUGCCUGCCAUUCGGUGGCGUUAGUGUAAAACCCUAAUGGCUAAUGCAUGACAGUCGAUG-ACGACGACUGGGGUGCCAGCGGCGGU------ -----.(((((((.((((..((((((.(((.(((....)))...)))..))))))....)))))))))))....-.((.((.((((....)))))).))..------ ( -34.20, z-score = -1.20, R) >droSec1.super_8 481955 95 - 3762037 -----UGAUUGUCUGUGCCUGCCAUUCGGUGGCGUUAGUGUAAAACCCUAAUGGCUAAUGCAUGACAGUCGAUG-ACGACGACUGGGGUGCCAGCGGCGGU------ -----.(((((((.((((..((((((.(((.(((....)))...)))..))))))....)))))))))))....-.((.((.((((....)))))).))..------ ( -34.20, z-score = -1.20, R) >droYak2.chrX 16800786 101 - 21770863 -----CGAUUGUCUGUGCCUGCCAUUCGAUGGCGUUAGUGUAAGACCCUAAUAGCUAAUGCAUGACAGUCGAUG-GCGAUGACUGGGGUGCCGGCAUCGGUGGCGGU -----..((((((...(((.(((((.((((.((((((((.((......))...))))))))......)))))))-))((((.((((....))))))))))))))))) ( -36.70, z-score = -0.93, R) >droEre2.scaffold_4690 8496573 95 - 18748788 -----CGAUUGUCUGUGCCUGCCAUUUGGUGGCGUUAGUGUAAAACCCUAAUAACUAAUGCAUGACGGUCUAUG-GCGAUGACUGGAGUGCCGGCGGCGGU------ -----..........((((.(((..(((((...(((((.(.....).))))).))))).((((..(((((....-.....)))))..)))).)))))))..------ ( -28.60, z-score = -0.12, R) >droGri2.scaffold_15081 3363766 101 + 4274704 CAUGCAUGUGGAAUCUAUUUACCAGUUGCCCGGGC--GUGUGCGGCGCCUGUCAAU-GUAUAUGGCCAAAUAUGCCCAAUGUGUAACGCCUGGUUUAUGAUGCU--- ((((((..(((((.....)).)))..)))((((((--(((..((((....)))...-((((((......)))))).....)..).))))))))...))).....--- ( -26.90, z-score = 1.42, R) >consensus _____UGAUUGUCUGUGCCUGCCAUUCGGUGGCGUUAGUGUAAAACCCUAAUGGCUAAUGCAUGACAGUCGAUG_ACGAUGACUGGGGUGCCAGCGGCGGU______ .....((((((((.((((..(((((...)))))((((((..............)))))))))))))))))).....((.((.((((....)))))).))........ (-23.64 = -21.56 + -2.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:59 2011