| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,152,838 – 18,152,942 |
| Length | 104 |
| Max. P | 0.999966 |

| Location | 18,152,838 – 18,152,942 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.91 |
| Shannon entropy | 0.66114 |
| G+C content | 0.47406 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.40 |
| Mean z-score | -4.58 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.24 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
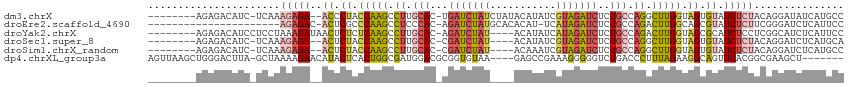
>dm3.chrX 18152838 104 + 22422827 --------AGAGACAUC-UCAAAGAGA--ACCCUACCAAGCCUUGCAC-UGAUCUAUCUAUACAUAUCGUAGAUCUCUGCCAGGCUUGGUAGUGUAUCUCUACAGGAUAUCAUGCC --------...((.(((-(...(((((--((.(((((((((((.(((.-.(((((((...........)))))))..))).))))))))))).)).)))))...)))).))..... ( -43.10, z-score = -6.23, R) >droEre2.scaffold_4690 8480911 91 + 18748788 ----------------------AGAGAC-ACUCGCCCAAGCCUCCCAC-AGAUCUAUGCACACAU-UCAUAGAUCUCUGCCAGACUUGGCAGCGUAUCUCUUCGGGAUCUCAUUCC ----------------------.((((.-....((....)).((((..-(((((((((.......-.)))))))))(((((......)))))...........))))))))..... ( -28.20, z-score = -3.42, R) >droYak2.chrX 16783544 103 + 21770863 --------AGAGACAUCCUCCUAAAGAUAACUCUCUCAAGCCUUGCAC-AGAUCUAU----ACAUAUCAUAGAUCUCUGCCAGACUUGGUAGCGCAUCUCCUCGGCAUCUCAUUCC --------.((((...((......((((..(.((..((((.((.(((.-((((((((----.......)))))))).))).)).))))..)).).))))....))..))))..... ( -25.80, z-score = -2.55, R) >droSec1.super_8 465743 100 + 3762037 --------AGAGACAUC-UCAAAGAGA--ACUCUACCAAGCCUUGCAC-CGAUCUAU----ACAUAUCGUAGAUCUCUGCCAGGCUUGGUAGUGUAUCUCUACAGGAUCUCAUGCA --------.((((..((-(...(((((--((.(((((((((((.(((.-.(((((((----.......)))))))..))).))))))))))).)).)))))...)))))))..... ( -45.20, z-score = -7.03, R) >droSim1.chrX_random 4718484 100 + 5698898 --------AGAGACAUC-UCAAAGAGA--ACUCUACCAAGCCUUGCAC-CGAUCUAU----ACAAAUCGUAGAUCUCUGCCAGGCUUGGUAGUGUAUCUCUACAGGAUCUCAUGCC --------.((((..((-(...(((((--((.(((((((((((.(((.-.(((((((----.......)))))))..))).))))))))))).)).)))))...)))))))..... ( -45.20, z-score = -7.38, R) >dp4.chrXL_group3a 123639 104 + 2690836 AGUUAAGCUGGGACUUA-GCUAAAAGAACAUACUCACUGGCGAUGGACGCGGUGUAA----GAGCCGAAAGGGGGUCUGACCCUUUAGAAGGCAGUUCACGGCGAAGCU------- ((((..(((((((((..-................(((((.((.....)))))))...----..(((.((((((.......))))))....)))))))).))))..))))------- ( -32.30, z-score = -0.87, R) >consensus ________AGAGACAUC_UCAAAGAGA__ACUCUACCAAGCCUUGCAC_CGAUCUAU____ACAUAUCAUAGAUCUCUGCCAGGCUUGGUAGCGUAUCUCUACAGGAUCUCAUGCC ......................(((((...(.((.(((((.((.(((...(((((((...........)))))))..))).)).))))).)).)..)))))............... (-14.62 = -15.52 + 0.90)

| Location | 18,152,838 – 18,152,942 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 63.91 |
| Shannon entropy | 0.66114 |
| G+C content | 0.47406 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -20.65 |
| Energy contribution | -22.49 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.28 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.33 |
| SVM RNA-class probability | 0.999966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18152838 104 - 22422827 GGCAUGAUAUCCUGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUAUGUAUAGAUAGAUCA-GUGCAAGGCUUGGUAGGGU--UCUCUUUGA-GAUGUCUCU-------- .....(((((((...(((((.((.(((((((((((.(((..(((((((..........).)))))).-.))).))))))))))).))--)))))..).-))))))...-------- ( -44.60, z-score = -4.82, R) >droEre2.scaffold_4690 8480911 91 - 18748788 GGAAUGAGAUCCCGAAGAGAUACGCUGCCAAGUCUGGCAGAGAUCUAUGA-AUGUGUGCAUAGAUCU-GUGGGAGGCUUGGGCGAGU-GUCUCU---------------------- (((......)))...((((((((..((((((((((..((.(((((((((.-.......)))))))))-.))..)))))).)))).))-))))))---------------------- ( -40.60, z-score = -4.67, R) >droYak2.chrX 16783544 103 - 21770863 GGAAUGAGAUGCCGAGGAGAUGCGCUACCAAGUCUGGCAGAGAUCUAUGAUAUGU----AUAGAUCU-GUGCAAGGCUUGAGAGAGUUAUCUUUAGGAGGAUGUCUCU-------- .....((((((((..((((((((.((..(((((((.(((.(((((((((.....)----))))))))-.))).)))))))..)).).)))))))....)).)))))).-------- ( -41.30, z-score = -4.40, R) >droSec1.super_8 465743 100 - 3762037 UGCAUGAGAUCCUGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUAUGU----AUAGAUCG-GUGCAAGGCUUGGUAGAGU--UCUCUUUGA-GAUGUCUCU-------- .....(((((.((..(((((.((.(((((((((((.(((..((((((........----.)))))).-.))).))))))))))).))--)))))...)-)..))))).-------- ( -43.40, z-score = -4.95, R) >droSim1.chrX_random 4718484 100 - 5698898 GGCAUGAGAUCCUGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUUUGU----AUAGAUCG-GUGCAAGGCUUGGUAGAGU--UCUCUUUGA-GAUGUCUCU-------- .....(((((.((..(((((.((.(((((((((((.(((..((((((........----.)))))).-.))).))))))))))).))--)))))...)-)..))))).-------- ( -43.40, z-score = -4.81, R) >dp4.chrXL_group3a 123639 104 - 2690836 -------AGCUUCGCCGUGAACUGCCUUCUAAAGGGUCAGACCCCCUUUCGGCUC----UUACACCGCGUCCAUCGCCAGUGAGUAUGUUCUUUUAGC-UAAGUCCCAGCUUAACU -------.(((.......((((.(((....((((((.......)))))).))).(----((((...(((.....)))..)))))...))))....)))-(((((....)))))... ( -23.60, z-score = -0.41, R) >consensus GGCAUGAGAUCCCGUAGAGAUACACUACCAAGCCUGGCAGAGAUCUACGAUAUGU____AUAGAUCG_GUGCAAGGCUUGGGAGAGU__UCUCUUUGA_GAUGUCUCU________ ...............(((((....(((((((((((.(((..((((((.............))))))...))).))))))))))).....)))))...................... (-20.65 = -22.49 + 1.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:58 2011