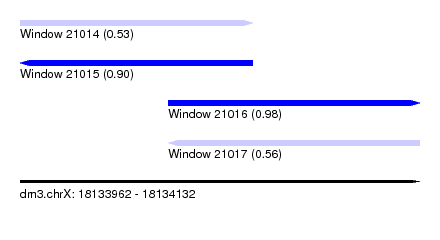
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,133,962 – 18,134,132 |
| Length | 170 |
| Max. P | 0.975612 |

| Location | 18,133,962 – 18,134,061 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.13 |
| Shannon entropy | 0.19002 |
| G+C content | 0.54026 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.92 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
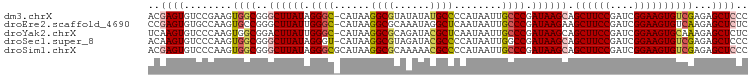
>dm3.chrX 18133962 99 + 22422827 ACGAGUGUCCGAAGUGGCGGGCUUAUAGGGC-CAUAAGGCGUAUAUAUGCCCCAUAAUUGCCCGAUAAGCAGCUUCCGAUCGGAAGUGUCGAGAGCUCCC ..((((.(((((........((((((.((((-.....(((((....)))))........)))).)))))).((((((....)))))).))).)))))).. ( -35.12, z-score = -1.86, R) >droEre2.scaffold_4690 8464385 99 + 18748788 CCGAGUGUGCCAAGUGCCGGGCUUAUUGGGC-CAUAAGGCGCAAAUAGGCUCAAUAAUUGCCCGAUAAGAAGCUUCCGAUCGGAAGUGUCAAGAGCUCUC ..((((...........((((((((((((((-(..............))))))))))..)))))....((.((((((....)))))).))....)))).. ( -36.34, z-score = -2.48, R) >droYak2.chrX 16763668 99 + 21770863 UCAAGUGUCCCAAGUGGCGGACUUAUUGGGC-CAUAAGGCGCAGAUACGCUCAAUAAUUGCCCGAUAAGCAGCUUCCGAUCGGAAGUGCAAAGAGCUCUC ......((((........))))(((((((((-.....((((......))))........)))))))))(((.(((((....))))))))........... ( -30.32, z-score = -1.09, R) >droSec1.super_8 446312 99 + 3762037 ACAAGUGUCCCAAGUGGCGGGCUUAUAGGGU-CAUAAGGCGUAGAUACGCCCCAUAAUUGGCCGAUAAGCAGCUUCCGAUCGGAAGUGUCGAGAGCUCCC ......(((......)))((((((((..(((-((...(((((....))))).......))))).))))))((((..(((((....).))))..)))))). ( -34.30, z-score = -1.72, R) >droSim1.chrX 13999005 100 + 17042790 ACGAGUGUCCCAAGUGGCGGGCUUAUAGGGCGCAUAAGGCGCAAAAACGCCCCAUAAUUGCCCGAUAAGCAGCUUCCGAUCGGAAGUGUCGAGAGCUCCC ..((((........((((..((((((.(((((.....((((......)))).......))))).)))))).((((((....))))))))))...)))).. ( -35.60, z-score = -1.65, R) >consensus ACGAGUGUCCCAAGUGGCGGGCUUAUAGGGC_CAUAAGGCGCAAAUACGCCCCAUAAUUGCCCGAUAAGCAGCUUCCGAUCGGAAGUGUCGAGAGCUCCC ..((((........((((..((((((.((((......((((......))))........)))).)))))).((((((....))))))))))...)))).. (-26.20 = -26.92 + 0.72)
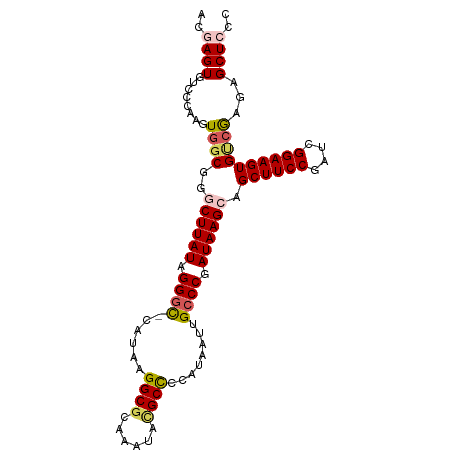
| Location | 18,133,962 – 18,134,061 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Shannon entropy | 0.19002 |
| G+C content | 0.54026 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -29.14 |
| Energy contribution | -30.38 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18133962 99 - 22422827 GGGAGCUCUCGACACUUCCGAUCGGAAGCUGCUUAUCGGGCAAUUAUGGGGCAUAUAUACGCCUUAUG-GCCCUAUAAGCCCGCCACUUCGGACACUCGU (((((((..((((......).)))..))))((((((.((((...((((((((........))))))))-)))).))))))(((......)))...))).. ( -35.80, z-score = -2.43, R) >droEre2.scaffold_4690 8464385 99 - 18748788 GAGAGCUCUUGACACUUCCGAUCGGAAGCUUCUUAUCGGGCAAUUAUUGAGCCUAUUUGCGCCUUAUG-GCCCAAUAAGCCCGGCACUUGGCACACUCGG (((.(((...((..(((((....)))))..))...((((((..((((((.((......))(((....)-)).)))))))))))).....)))...))).. ( -30.20, z-score = -1.52, R) >droYak2.chrX 16763668 99 - 21770863 GAGAGCUCUUUGCACUUCCGAUCGGAAGCUGCUUAUCGGGCAAUUAUUGAGCGUAUCUGCGCCUUAUG-GCCCAAUAAGUCCGCCACUUGGGACACUUGA (((.((((...((((((((....))))).))).....))))..((((((.(((....)))(((....)-)).))))))((((........)))).))).. ( -30.60, z-score = -1.10, R) >droSec1.super_8 446312 99 - 3762037 GGGAGCUCUCGACACUUCCGAUCGGAAGCUGCUUAUCGGCCAAUUAUGGGGCGUAUCUACGCCUUAUG-ACCCUAUAAGCCCGCCACUUGGGACACUUGU ((.((((..((((......).)))..))))((((((.((....(((((((((((....))))))))))-).)).)))))))).((....))......... ( -32.60, z-score = -1.61, R) >droSim1.chrX 13999005 100 - 17042790 GGGAGCUCUCGACACUUCCGAUCGGAAGCUGCUUAUCGGGCAAUUAUGGGGCGUUUUUGCGCCUUAUGCGCCCUAUAAGCCCGCCACUUGGGACACUCGU (((.(.((((((..(((((....)))))..((((((.((((...((((((((((....)))))))))).)))).)))))).......))))))).))).. ( -41.10, z-score = -2.87, R) >consensus GGGAGCUCUCGACACUUCCGAUCGGAAGCUGCUUAUCGGGCAAUUAUGGGGCGUAUCUGCGCCUUAUG_GCCCUAUAAGCCCGCCACUUGGGACACUCGU (((.(.((((((..(((((....)))))..((((((.((((...((((((((((....)))))))))).)))).)))))).......))))))).))).. (-29.14 = -30.38 + 1.24)

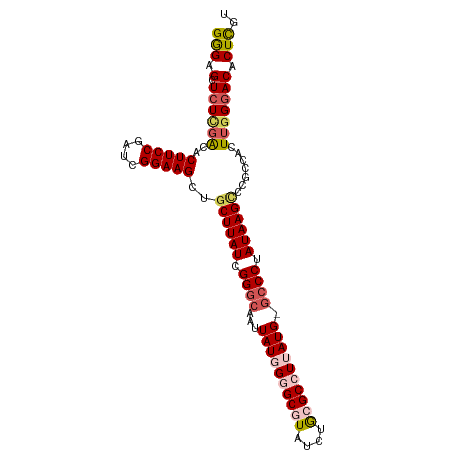
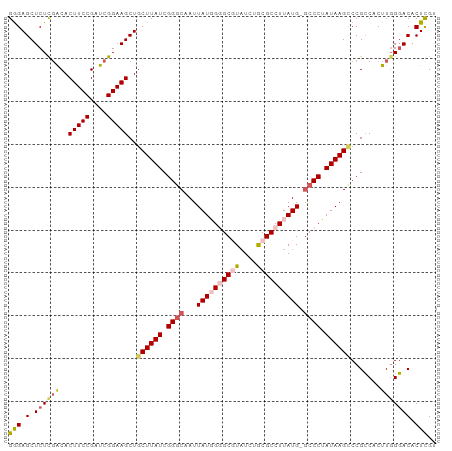
| Location | 18,134,025 – 18,134,132 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.23 |
| Shannon entropy | 0.56776 |
| G+C content | 0.49435 |
| Mean single sequence MFE | -35.49 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18134025 107 + 22422827 AUAAGCAGCUUCCGAUCGGAAGUGUCGA---GAGCUCCCAUCGAACCGAUCGGCUGGGCUAUCAGUUCGCUAUCGUCGAUGGUGGAC---ACAAGCCUCAACUGACUGAUUAU--- ...(((((...((((((((.....((((---.........)))).)))))))).((((((....((((((((((...))))))))))---...)))).)).))).))......--- ( -36.00, z-score = -1.36, R) >droAna3.scaffold_13248 3732951 106 - 4840945 AUAAGCGUGUUCCGAUCGGAAGUUGGAAACAGAAACGUUUCGAAAACGUUUAGUCAAGUGGGCCAUAAA-UAUUGCCAGAGAUAGGGCUAAUAAGCC--AACUUCCGAU------- ..............((((((((((((......(((((((.....)))))))((((.....(((.((...-.)).)))........))))......))--))))))))))------- ( -30.52, z-score = -2.70, R) >droEre2.scaffold_4690 8464448 110 + 18748788 AUAAGAAGCUUCCGAUCGGAAGUGUCAA---GAGCUCUCGUCGAACCGAUCGGCUGGGCUAACAGUUCGCUAUCGGCGAUGGUGGCA---ACAAGCCCCAACCCUUUGAAUUUUCC ....((.((((((....)))))).))..---.(((((..(((((.....))))).)))))......((((.....))))(((.(((.---....))))))................ ( -34.50, z-score = -1.03, R) >droYak2.chrX 16763731 110 + 21770863 AUAAGCAGCUUCCGAUCGGAAGUGCAAA---GAGCUCUCAGCGAACCGAUCGACUGCGUUAUCAGUUCGCUAUCGUCGAUGGUGGAA---ACAAGCCUCAACUGCUAAAGAAUUAU ....(((.(((((....))))))))...---.(((....(((((((.(((.(((...)))))).)))))))...((.((.((((...---.)..))))).)).))).......... ( -31.20, z-score = -1.00, R) >droSec1.super_8 446375 110 + 3762037 AUAAGCAGCUUCCGAUCGGAAGUGUCGA---GAGCUCCCAUCGAACCGAUCGGCUGGGCUAUCAGUUUGCUAUCGUCGAUGGUGGACCCAAUCAGUCUCAACUGACUGAUUAU--- ......(((((((((((((.....((((---.........)))).))))))))..)))))....(((..(((((...)))))..)))..((((((((......))))))))..--- ( -40.50, z-score = -2.63, R) >droSim1.chrX 13999069 110 + 17042790 AUAAGCAGCUUCCGAUCGGAAGUGUCGA---GAGCUCCCAUCGAACCGAUCGGCUGGGCUAUCAGUUCGCUAUCGUCGAUGGCGGACUGUAUCACUCUUAACUGACUGAUUAU--- ......(((((((((((((.....((((---.........)))).))))))))..)))))..((((((((((((...)))))))))))).((((.((......)).))))...--- ( -40.20, z-score = -2.45, R) >consensus AUAAGCAGCUUCCGAUCGGAAGUGUCAA___GAGCUCCCAUCGAACCGAUCGGCUGGGCUAUCAGUUCGCUAUCGUCGAUGGUGGAC___ACAAGCCUCAACUGACUGAUUAU___ .......((((((....))))))...........((.(((((((..((((.(((.(((((...))))))))))))))))))).))............................... (-18.20 = -18.27 + 0.06)

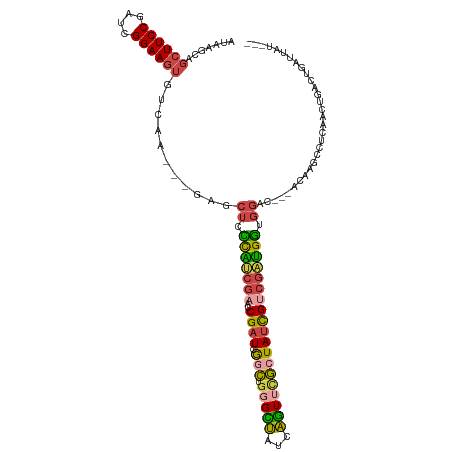
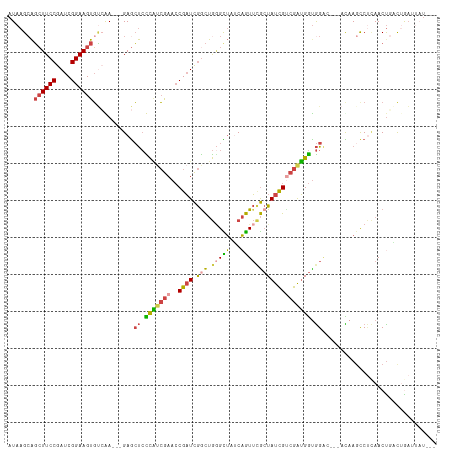
| Location | 18,134,025 – 18,134,132 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.23 |
| Shannon entropy | 0.56776 |
| G+C content | 0.49435 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -12.48 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18134025 107 - 22422827 ---AUAAUCAGUCAGUUGAGGCUUGU---GUCCACCAUCGACGAUAGCGAACUGAUAGCCCAGCCGAUCGGUUCGAUGGGAGCUC---UCGACACUUCCGAUCGGAAGCUGCUUAU ---((((.(((...((((((((((..---.....((((((.(....))((((((((.((...))..))))))))))))))))).)---))))).(((((....)))))))).)))) ( -34.91, z-score = -1.11, R) >droAna3.scaffold_13248 3732951 106 + 4840945 -------AUCGGAAGUU--GGCUUAUUAGCCCUAUCUCUGGCAAUA-UUUAUGGCCCACUUGACUAAACGUUUUCGAAACGUUUCUGUUUCCAACUUCCGAUCGGAACACGCUUAU -------((((((((((--((.......(((........)))....-..............(((.(((((((.....)))))))..))).)))))))))))).............. ( -27.80, z-score = -2.76, R) >droEre2.scaffold_4690 8464448 110 - 18748788 GGAAAAUUCAAAGGGUUGGGGCUUGU---UGCCACCAUCGCCGAUAGCGAACUGUUAGCCCAGCCGAUCGGUUCGACGAGAGCUC---UUGACACUUCCGAUCGGAAGCUUCUUAU ((((..(((...((((((((((....---.))).)).((((.....))))......)))))...(((((((.....((((....)---)))......))))))))))..))))... ( -37.70, z-score = -1.53, R) >droYak2.chrX 16763731 110 - 21770863 AUAAUUCUUUAGCAGUUGAGGCUUGU---UUCCACCAUCGACGAUAGCGAACUGAUAACGCAGUCGAUCGGUUCGCUGAGAGCUC---UUUGCACUUCCGAUCGGAAGCUGCUUAU ..........(((.(((((((.....---.....)).)))))..((((((((((((..........))))))))))))...))).---...((((((((....))))).))).... ( -34.30, z-score = -1.61, R) >droSec1.super_8 446375 110 - 3762037 ---AUAAUCAGUCAGUUGAGACUGAUUGGGUCCACCAUCGACGAUAGCAAACUGAUAGCCCAGCCGAUCGGUUCGAUGGGAGCUC---UCGACACUUCCGAUCGGAAGCUGCUUAU ---..((((((((......))))))))(((..(.(((((((((((......(((......)))...))))..))))))))..)))---..(.(((((((....))))).))).... ( -35.80, z-score = -1.47, R) >droSim1.chrX 13999069 110 - 17042790 ---AUAAUCAGUCAGUUAAGAGUGAUACAGUCCGCCAUCGACGAUAGCGAACUGAUAGCCCAGCCGAUCGGUUCGAUGGGAGCUC---UCGACACUUCCGAUCGGAAGCUGCUUAU ---((((.(((...((..(((((....((((.(((.(((...))).))).))))....((((..(((.....))).)))).))))---)..)).(((((....)))))))).)))) ( -36.00, z-score = -1.73, R) >consensus ___AUAAUCAGUCAGUUGAGGCUUAU___GUCCACCAUCGACGAUAGCGAACUGAUAGCCCAGCCGAUCGGUUCGAUGAGAGCUC___UCGACACUUCCGAUCGGAAGCUGCUUAU ................((((..............(((((((((((......(((......)))...))))..)))))))...............(((((....)))))...)))). (-12.48 = -13.15 + 0.67)

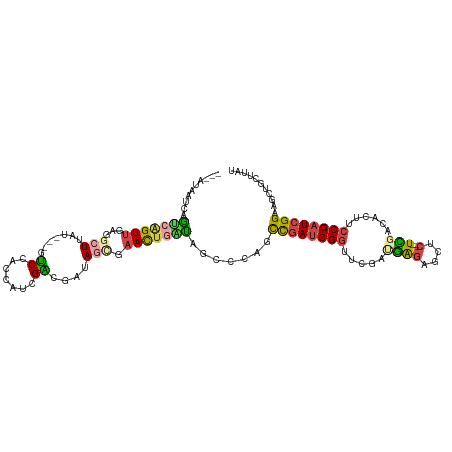
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:53 2011