| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,132,500 – 18,132,650 |
| Length | 150 |
| Max. P | 0.987949 |

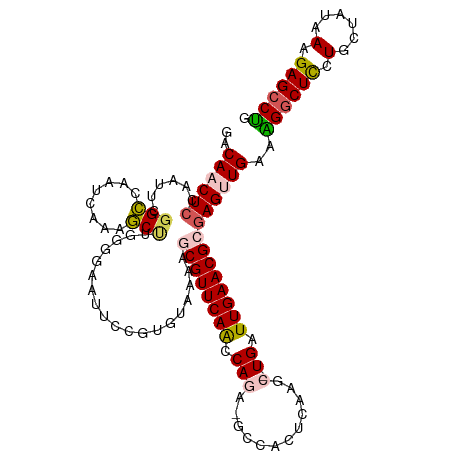
| Location | 18,132,500 – 18,132,614 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.31141 |
| G+C content | 0.47039 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
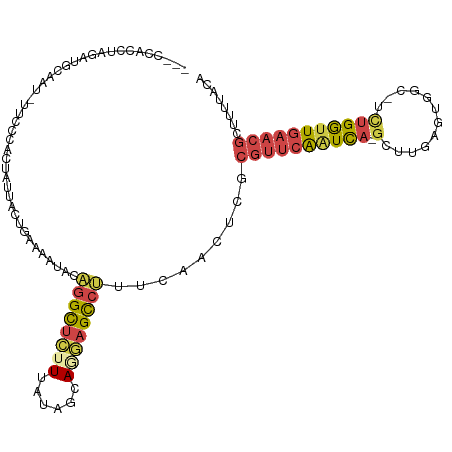
>dm3.chrX 18132500 114 + 22422827 GACAACUCGAAUUCGGCCAAUCAAAGCUUGGGGAAUUCCGUGUAAAACCGUUCAGCCAGA-GCCACUCAACCCUGAUUGAACGCGAGUUGAAAGGCUUCUGCUAUAAAGAGCCUG ..((((((((((((..((((.......)))).))))))..........((((((..(((.-(........).)))..)))))).))))))..(((((.((.......))))))). ( -33.30, z-score = -1.95, R) >droSim1.chrX 13997513 113 + 17042790 GACAACUCCAAUUUGGCCAAUCAAAGCUUGGGGAGUUCCGUGUAAAAGCGUUCAACCAGA-GCCACUCGAGC-UGAUUGAACGCGAGUUGAAAGGCUCCUGCUAUAAAGAGCCUG ..((((((.....((..((((((..(((((((..(((((((......)))........))-))..)))))))-))))))..)).))))))..((((((.(......).)))))). ( -36.40, z-score = -2.20, R) >droSec1.super_8 444830 113 + 3762037 GACAACUCCAAUUCGGCCAAUCGAAGCUUGGGGAAUUCCGUGUAAAAGCGUUCAACCAGA-GCCACUCAAGC-UGAUUGAACGCGAGUUGAAAGACUCCUGCUAUAAAGAGCCUG .....(((((((((((....)))))..))))))..(((..((((...((((((((.(((.-..........)-)).))))))))(((((....)))))....))))..))).... ( -31.80, z-score = -1.74, R) >droYak2.chrX 16762195 104 + 21770863 AACA-CUCCAAUUUGGUCAAUCAGAAC---------UCCGUGAAAUUGCGUUCAACCAAAAGCAGUUCAAUC-UGAUUGAACGCAAGUUGAAGGGCUCUUGCUAUAAAGAGCCUG ....-((.(((((((((((((((((..---------....((((.((((............)))))))).))-)))))).)).))))))).))(((((((......))))))).. ( -35.20, z-score = -4.50, R) >droEre2.scaffold_4690 8462927 112 + 18748788 GACA-CUCAAAUUCGGUCAAAGAAAACCUGGGGAAUUCCGUGAAAACACGUUCAACCAAA-GCCAUGCAAGC-UGAUUGAACGAGAGUUGAAUGGCUCGUGCUAUAAAGAGCCAG (((.-(((...(((((((..........(((((....))(((....)))......))).(-((.......))-)))))))).))).)))...((((((..........)))))). ( -27.50, z-score = -1.01, R) >consensus GACAACUCCAAUUCGGCCAAUCAAAGCUUGGGGAAUUCCGUGUAAAAGCGUUCAACCAGA_GCCACUCAAGC_UGAUUGAACGCGAGUUGAAAGGCUCCUGCUAUAAAGAGCCUG ..((((((......(((........)))...................((((((((.((...((.......)).)).))))))))))))))..((((((.(......).)))))). (-21.42 = -21.86 + 0.44)

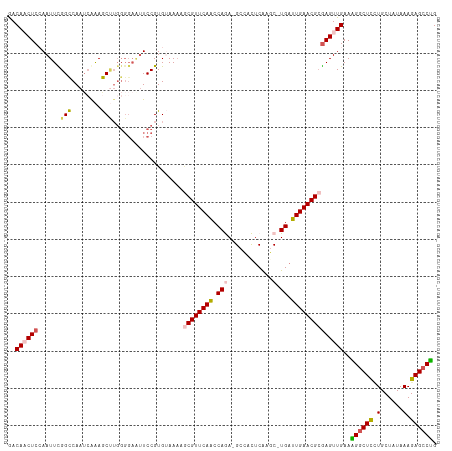
| Location | 18,132,500 – 18,132,614 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.31141 |
| G+C content | 0.47039 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -27.40 |
| Energy contribution | -28.44 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18132500 114 - 22422827 CAGGCUCUUUAUAGCAGAAGCCUUUCAACUCGCGUUCAAUCAGGGUUGAGUGGC-UCUGGCUGAACGGUUUUACACGGAAUUCCCCAAGCUUUGAUUGGCCGAAUUCGAGUUGUC .(((((((.......)).)))))..((((((.((((((..(((((((....)))-))))..))))))..........((((((.((((.......))))..)))))))))))).. ( -40.10, z-score = -2.58, R) >droSim1.chrX 13997513 113 - 17042790 CAGGCUCUUUAUAGCAGGAGCCUUUCAACUCGCGUUCAAUCA-GCUCGAGUGGC-UCUGGUUGAACGCUUUUACACGGAACUCCCCAAGCUUUGAUUGGCCAAAUUGGAGUUGUC .((((((((......))))))))..(((((((((((((((((-(..(.....).-.))))))))))))........((......))..(((......))).......)))))).. ( -38.80, z-score = -2.45, R) >droSec1.super_8 444830 113 - 3762037 CAGGCUCUUUAUAGCAGGAGUCUUUCAACUCGCGUUCAAUCA-GCUUGAGUGGC-UCUGGUUGAACGCUUUUACACGGAAUUCCCCAAGCUUCGAUUGGCCGAAUUGGAGUUGUC .((((((((......))))))))..(((((((((((((((((-(((.....)))-..)))))))))))..........(((((.((((.......))))..))))).)))))).. ( -38.50, z-score = -2.46, R) >droYak2.chrX 16762195 104 - 21770863 CAGGCUCUUUAUAGCAAGAGCCCUUCAACUUGCGUUCAAUCA-GAUUGAACUGCUUUUGGUUGAACGCAAUUUCACGGA---------GUUCUGAUUGACCAAAUUGGAG-UGUU ..(((((((......)))))))((((((.(((((((((((((-((..........))))))))))))))).....(((.---------...)))..........))))))-.... ( -35.00, z-score = -3.35, R) >droEre2.scaffold_4690 8462927 112 - 18748788 CUGGCUCUUUAUAGCACGAGCCAUUCAACUCUCGUUCAAUCA-GCUUGCAUGGC-UUUGGUUGAACGUGUUUUCACGGAAUUCCCCAGGUUUUCUUUGACCGAAUUUGAG-UGUC .((((((..........))))))....((.((((((((((((-(((.....)))-..)))))))))(((....))).((((((....((((......)))))))))))))-.)). ( -32.90, z-score = -2.25, R) >consensus CAGGCUCUUUAUAGCAGGAGCCUUUCAACUCGCGUUCAAUCA_GCUUGAGUGGC_UCUGGUUGAACGCUUUUACACGGAAUUCCCCAAGCUUUGAUUGGCCGAAUUGGAGUUGUC .((((((((......))))))))..(((((((((((((((((.(((.....)))...)))))))))))....................(((......))).......)))))).. (-27.40 = -28.44 + 1.04)

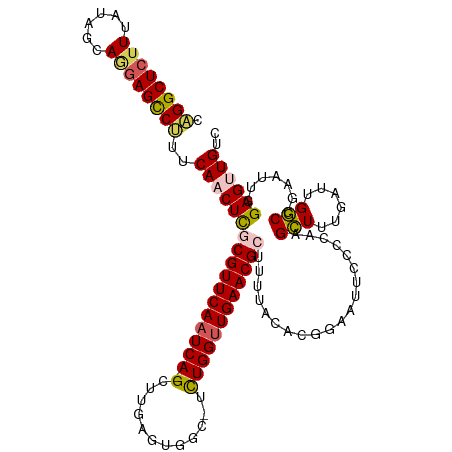
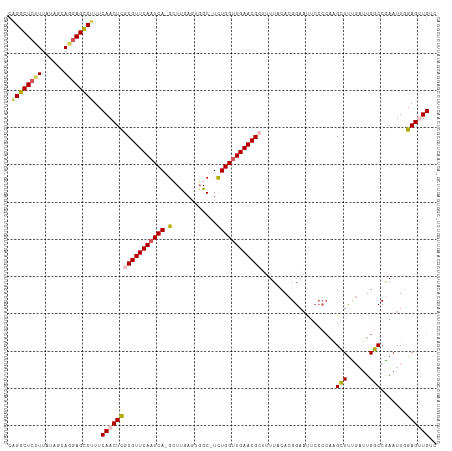
| Location | 18,132,540 – 18,132,650 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.56 |
| Shannon entropy | 0.52935 |
| G+C content | 0.43534 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -12.40 |
| Energy contribution | -13.16 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.941852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
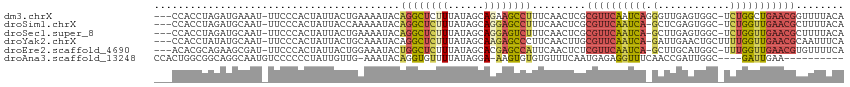
>dm3.chrX 18132540 110 - 22422827 ---CCACCUAGAUGAAAU-UUCCCACUAUUACUGAAAAUACAGGCUCUUUAUAGCAGAAGCCUUUCAACUCGCGUUCAAUCAGGGUUGAGUGGC-UCUGGCUGAACGGUUUUACA ---.........((((((-.............((((.....(((((((.......)).))))))))).....((((((..(((((((....)))-))))..)))))))))))).. ( -26.00, z-score = -0.54, R) >droSim1.chrX 13997553 109 - 17042790 ---CCACCUAGAUGCAAU-UUCCCACUAUUACCAAAAAUACAGGCUCUUUAUAGCAGGAGCCUUUCAACUCGCGUUCAAUCA-GCUCGAGUGGC-UCUGGUUGAACGCUUUUACA ---...............-......................((((((((......))))))))........(((((((((((-(..(.....).-.))))))))))))....... ( -28.40, z-score = -2.49, R) >droSec1.super_8 444870 109 - 3762037 ---CCACCUAGAUGCAAU-UUCCCACUAUUACUGAAAAUACAGGCUCUUUAUAGCAGGAGUCUUUCAACUCGCGUUCAAUCA-GCUUGAGUGGC-UCUGGUUGAACGCUUUUACA ---...............-.............((((.....((((((((......))))))))))))....(((((((((((-(((.....)))-..)))))))))))....... ( -27.00, z-score = -1.52, R) >droYak2.chrX 16762225 110 - 21770863 ---CCACCUAUAUGCAAU-UUCCCACUAUUACUGCAAAUACAGGCUCUUUAUAGCAAGAGCCCUUCAACUUGCGUUCAAUCA-GAUUGAACUGCUUUUGGUUGAACGCAAUUUCA ---.....(((.((((..-.............)))).)))..(((((((......))))))).......(((((((((((((-((..........)))))))))))))))..... ( -30.86, z-score = -4.63, R) >droEre2.scaffold_4690 8462966 109 - 18748788 ---ACACGCAGAAGCGAU-UUCCCACUAUUACUGGAAAUACUGGCUCUUUAUAGCACGAGCCAUUCAACUCUCGUUCAAUCA-GCUUGCAUGGC-UUUGGUUGAACGUGUUUUCA ---...(((....)))..-..............((((((((((((((..........))))))..........(((((((((-(((.....)))-..))))))))))))))))). ( -32.40, z-score = -3.21, R) >droAna3.scaffold_13248 3728206 99 + 4840945 CCACUGGCGGCAGGCAAUGUCCCCCCUAUUGUUG-AAAUACAGGUGUUUUAUAGGA-AAGUGUGUGUUUCAAUGAGAGGUUUCAACCGAUUGGC----GAUUGAA---------- .....((.((((.....)))))).(((.(..(((-(((((((.(..(((.....))-)..).))))))))))..).))).(((((.((.....)----).)))))---------- ( -25.90, z-score = -0.41, R) >consensus ___CCACCUAGAUGCAAU_UUCCCACUAUUACUGAAAAUACAGGCUCUUUAUAGCAGGAGCCUUUCAACUCGCGUUCAAUCA_GCUUGAGUGGC_UCUGGUUGAACGCUUUUACA .........................................((((((((......)))))))).........((((((((((...............))))))))))........ (-12.40 = -13.16 + 0.76)
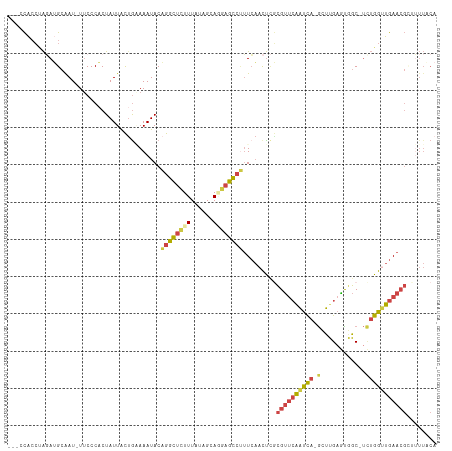
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:48 2011