| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,131,028 – 18,131,133 |
| Length | 105 |
| Max. P | 0.738544 |

| Location | 18,131,028 – 18,131,133 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.91 |
| Shannon entropy | 0.23657 |
| G+C content | 0.45194 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
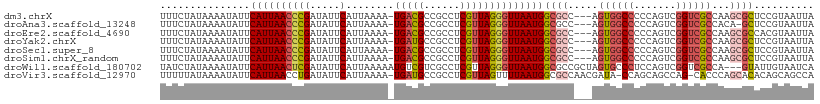
>dm3.chrX 18131028 105 + 22422827 UUUCUAUAAAAUAUUCAUUAACCCGAUAUUCAUUAAAA-UGACGCCGCCUCGUUAGGGUUAAUGGCGCC---AGUGGCCCCCAGUCGGUCGCCAAGCGCUCCGUAAUUA ...............((((((((((.....).......-(((((......))))))))))))))((((.---..((((..((....))..)))).)))).......... ( -30.70, z-score = -2.59, R) >droAna3.scaffold_13248 3726123 104 - 4840945 UUUCUAUAAAAUAUUCAUUAACCCGAUAUUCAUUAAAA-UGACGCCGCCUCGUUAGGGUUAAUGGCGCC---AGUGGCCCCCAGUCGGUCGCCACA-GCUCCGUAAUUA ................(((((((((.....).......-(((((......)))))))))))))((.((.---.(((((..((....))..))))).-)).))....... ( -28.10, z-score = -2.17, R) >droEre2.scaffold_4690 8461584 105 + 18748788 UUUCUAUAAAAUAUUCAUUAACCCGAUAUUCAUUAAAA-UGACGCCGCCUCGUUAGGGUUAAUGGCGCC---AGUGGCCCCCAGUCGGUCGCCAAGCGCCACGUAAUUA ................(((((((((.....).......-(((((......)))))))))))))(((((.---..((((..((....))..)))).)))))......... ( -31.50, z-score = -2.79, R) >droYak2.chrX 16760588 105 + 21770863 UUUCUAUAAAAUAUUCAUUAACCCGAUAUUCAUUAAAA-UGAUGCCGCCUCGUUAGGGUUAAUGGCGCC---AGUGGCCCCCAGUCGGUCGCCAAGCGCUCCGUAAUUA ...............((((((((((.....).....((-(((.......))))).)))))))))((((.---..((((..((....))..)))).)))).......... ( -29.10, z-score = -2.09, R) >droSec1.super_8 443338 105 + 3762037 UUUCUAUAAAAUAUUCAUUAACCCGAUAUUCAUUAAAA-UGACGCCGCCUCGUUAGGGUUAAUGGCGCC---AGUGGCCCCCAGUCGGUCGCCAAGCGCUCCGUAAUUA ...............((((((((((.....).......-(((((......))))))))))))))((((.---..((((..((....))..)))).)))).......... ( -30.70, z-score = -2.59, R) >droSim1.chrX_random 228913 105 + 5698898 UUUCUAUAAAAUAUUCAUUAACCCGAUAUUCAUUAAAA-UGACGCCGCCUCGUUAGGGUUAAUGGCGCC---AGUGGCCCCCAGUCGGUCGCCAAGCGCUCCGUAAUUA ...............((((((((((.....).......-(((((......))))))))))))))((((.---..((((..((....))..)))).)))).......... ( -30.70, z-score = -2.59, R) >droWil1.scaffold_180702 1338878 106 + 4511350 UAUCUAUAAAAUAUUCAUUAACUCGAUAUUCAUUAAAAAUGUCGUCGCCUCGUUAGGGUUAAUGGCGCCGCUAGUGCCCUCCAGUCGGUCGCCA---GUAUUGUAAUCA .........((((((.((((((((((((((.......))))))(......)....))))))))(((((((((.(......).)).))).)))))---)))))....... ( -22.90, z-score = -1.58, R) >droVir3.scaffold_12970 2853494 106 + 11907090 UUUUUAUAAAAUAUUCAUUAACCUGAUAUUCAUUAAAA-UGAUGCCGCCUCGUUAGUUUUAAUGGCGCCAACGAUA-CCAGCAGCCAG-CACCCAGCACACAGCAGCCA ..............(((((....((.....))....))-))).((.((..............((((((........-...)).))))(-(.....)).....)).)).. ( -12.30, z-score = 1.23, R) >consensus UUUCUAUAAAAUAUUCAUUAACCCGAUAUUCAUUAAAA_UGACGCCGCCUCGUUAGGGUUAAUGGCGCC___AGUGGCCCCCAGUCGGUCGCCAAGCGCUCCGUAAUUA ...............((((((((((.....)........(((((......))))))))))))))((((.....((((((.......))))))...)))).......... (-19.93 = -20.85 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:46 2011