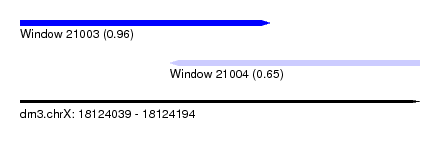
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,124,039 – 18,124,194 |
| Length | 155 |
| Max. P | 0.962506 |

| Location | 18,124,039 – 18,124,136 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.12 |
| Shannon entropy | 0.47854 |
| G+C content | 0.55525 |
| Mean single sequence MFE | -35.89 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.60 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
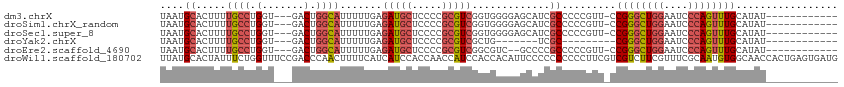
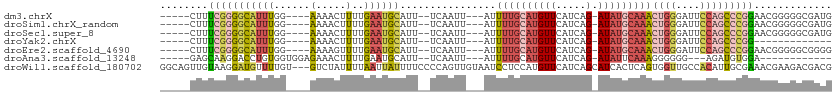
>dm3.chrX 18124039 97 + 22422827 UAAUGCACUUUUGCCUGGU---GACUGGCAUUUUUGAGAUGCUCCCCGCGUCGGUGGGGAGCAUCGCCCCCGUU-CCGGGCUGGAAUCCCAGUUUGCAUAU------------ ..(((((....((((.(..---..).)))).......((((((((((((....))))))))))))((((.....-..))))(((....)))...)))))..------------ ( -45.90, z-score = -4.02, R) >droSim1.chrX_random 220663 97 + 5698898 UAAUGCACUUUUGCCUGGU---GACUGGCAUUUUUGAGAUGCUCCCCGCGUCGGUGGGGAGCAUCGCCCCCGUU-CCGGGCUGGAAUCCCAGUUUGCAUAU------------ ..(((((....((((.(..---..).)))).......((((((((((((....))))))))))))((((.....-..))))(((....)))...)))))..------------ ( -45.90, z-score = -4.02, R) >droSec1.super_8 436583 97 + 3762037 UAAUGCACUUUUGCCUGGU---GACUGGCAUUUUUGAGAUGCUCCCCGCGUCGGUGGGGAGCAUCGCCCCCGUU-CCGGGCUGGAAUCCCAGUUUGCAUAU------------ ..(((((....((((.(..---..).)))).......((((((((((((....))))))))))))((((.....-..))))(((....)))...)))))..------------ ( -45.90, z-score = -4.02, R) >droYak2.chrX 16752912 82 + 21770863 UAAUGCACUUUUGCCUGGU---GACUGGCAUUUUUGAGAUGCUCCCCGCGUCGCUG-------UCGC---------CGGGCUGGAAUCCCAGUUUGCAUAU------------ ..(((((.....(((((((---(((.(((........(((((.....)))))))))-------))))---------)))))(((....)))...)))))..------------ ( -32.50, z-score = -3.06, R) >droEre2.scaffold_4690 8452695 95 + 18748788 UAAUGCACUUUUGCCUGGU---GACUGGCAUUUUUGAGAUGCUCCCCGCGUCGGCGUC--GCCCCGCCCCCGUU-CCGGGCUGGAAUCCCAGUUUGCAUAU------------ ..(((((.....((((((.---(((.((.........(((((.....)))))((((..--....)))))).)))-))))))(((....)))...)))))..------------ ( -33.50, z-score = -1.60, R) >droWil1.scaffold_180702 1330804 113 + 4511350 UUAUGCACUAUUUCUGGUUUCCGACCCAACUUUUCAUCAUCCACCAACCAUCCACCACAUUCCCCCCCCCCUUCGUCGUCUUCGUUUCGCAAUGUGGCAACCACUGAGUGAUG .....((((.....(((((...((.........))...................(((((((............((.((....))...)).))))))).)))))...))))... ( -11.62, z-score = 0.76, R) >consensus UAAUGCACUUUUGCCUGGU___GACUGGCAUUUUUGAGAUGCUCCCCGCGUCGGUGGGGAGCAUCGCCCCCGUU_CCGGGCUGGAAUCCCAGUUUGCAUAU____________ ..((((.....((((.(.......).)))).......(((((.....))))).........................(((((((....))))))))))).............. (-14.57 = -15.60 + 1.03)
| Location | 18,124,097 – 18,124,194 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.42 |
| Shannon entropy | 0.52146 |
| G+C content | 0.43251 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -11.57 |
| Energy contribution | -13.07 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
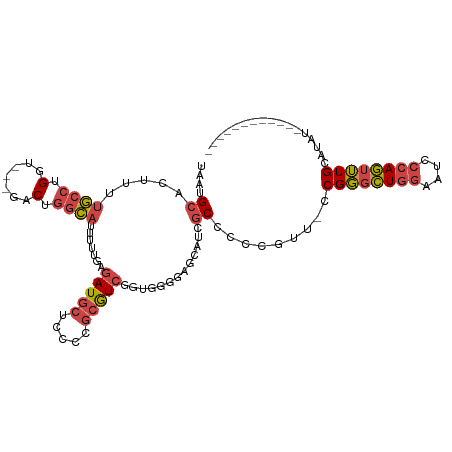
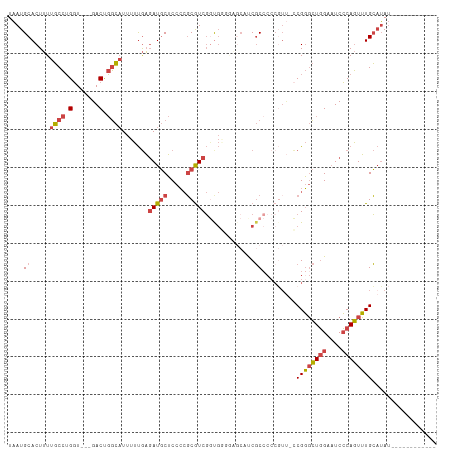
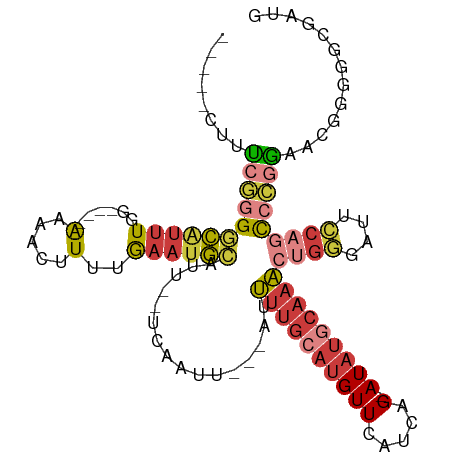
>dm3.chrX 18124097 97 - 22422827 -----CUUUCGGGGCAUUUGG----AAAACUUUUGAAUGCAUU--UCAAUU---AUUUUGCAUGUUCAUCAG-AUAUGCAAACUGGGAUUCCAGCCCGGAACGGGGGCGAUG -----.((((((((((((..(----(.....))..)))))...--......---..((((((((((.....)-)))))))))((((....)))))))))))........... ( -30.40, z-score = -2.52, R) >droSim1.chrX_random 220721 97 - 5698898 -----CUUUCGGGGCAUUUGG----AAAACUUUUGAAUGCAUU--UCAAUU---AUUUUGCAUGUUCAUCAG-AUAUGCAAACUGGGAUUCCAGCCCGGAACGGGGGCGAUG -----.((((((((((((..(----(.....))..)))))...--......---..((((((((((.....)-)))))))))((((....)))))))))))........... ( -30.40, z-score = -2.52, R) >droSec1.super_8 436641 97 - 3762037 -----CUUUCGGGGCAUUUGG----AAAACUUUUGAAUGCAUU--UCAAUU---AUUUUGCAUGUUCAUCAG-AUAUGCAAACUGGGAUUCCAGCCCGGAACGGGGGCGAUG -----.((((((((((((..(----(.....))..)))))...--......---..((((((((((.....)-)))))))))((((....)))))))))))........... ( -30.40, z-score = -2.52, R) >droYak2.chrX 16752968 84 - 21770863 -----CUUUCGGGGCAUUUGG----AAAACUUUUGAAUGCAUU--UCAAUU---AUUUUGCAUGUUCAUCAG-AUAUGCAAACUGGGAUUCCAGCCCGG------------- -----....(((((((((..(----(.....))..)))))...--......---..((((((((((.....)-)))))))))((((....)))))))).------------- ( -28.00, z-score = -3.60, R) >droEre2.scaffold_4690 8452751 97 - 18748788 -----CUUUCGGGGCAUUUGG----AAAAGUUUUGAAUGCAUU--UCAAUU---AUUUUGCAUGUUCAUCAG-AUAUGCAAACUGGGAUUCCAGCCCGGAACGGGGGCGGGG -----.(..(((.(((((..(----(.....))..)))))...--......---..((((((((((.....)-))))))))))))..)..((.((((.......)))).)). ( -31.50, z-score = -2.61, R) >droAna3.scaffold_13248 3716032 86 + 4840945 -----GAGCAAGGACCUGUGGUGGAGAAACUUUUGAAUGCAUU--UCAAUU---AUUUUGCAUGUUCAUCAG-AUAUUCAAAGGGGGG---AGAUGUGGA------------ -----..(((....(((.(..((((....((..((((((((..--......---....)))))..)))..))-...))))..).))).---...)))...------------ ( -15.30, z-score = 1.19, R) >droWil1.scaffold_180702 1330877 109 - 4511350 GGCAGUUGUAAGGAUGUUUUGU---GUCUAUUUUAAUUAUUUUCCCCAGUUGUAAUCCUCCAUGUUCAUCAGCAUCACUCAGUGGUUGCCACAUUGCGAAACGAAGACGACG ....(((((.....((((((((---.......................((((.....(.....).....))))........((((...))))...))))))))...))))). ( -17.30, z-score = 1.47, R) >consensus _____CUUUCGGGGCAUUUGG____AAAACUUUUGAAUGCAUU__UCAAUU___AUUUUGCAUGUUCAUCAG_AUAUGCAAACUGGGAUUCCAGCCCGGAACGGGGGCGAUG ........((((((((((..(...........)..)))))................(((((((((.(....).)))))))))((((....)))))))))............. (-11.57 = -13.07 + 1.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:42 2011