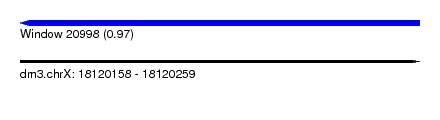
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,120,158 – 18,120,259 |
| Length | 101 |
| Max. P | 0.969951 |

| Location | 18,120,158 – 18,120,259 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 61.61 |
| Shannon entropy | 0.77286 |
| G+C content | 0.45148 |
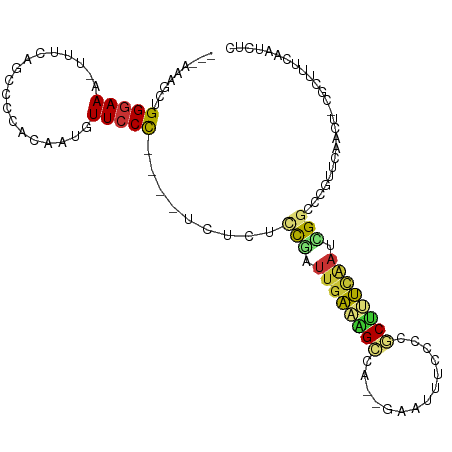
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -7.94 |
| Energy contribution | -7.61 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18120158 101 - 22422827 ---AAAGCUGGGAAAUUUUUAGCCCCAAAAUGUUCCC----UCUAUUCGAUUGAAAGCCAACGAAUUUCCCCGCUUUCAAUCGGCGCGUUCAACU-CGCUUUCAAUAUC ---...((.(((((..((((......))))..)))))----......(((((((((((..............)))))))))))))(((.......-))).......... ( -25.34, z-score = -2.82, R) >droSim1.chrX_random 4682677 100 - 5698898 ---AAAGCUGGGAAA-UUUCAGCCCCACAAUGUUGCC----UCUGUUCGAUUGAAAGCCAACGAAUUUCCCCGCUUUCAAUCGGCCCGUUCAACU-CGCUUUCAAUCUC ---(((((((((...-.......))))....((((..----.(.(..(((((((((((..............)))))))))))..).)..)))).-.)))))....... ( -26.14, z-score = -2.78, R) >droSec1.super_8 432775 103 - 3762037 GUAAAAGCUGGGAAA-UUUCAGCCCCACAAUGUUCCC----UCUGUUCGAUUGAAAGCCAACGAAUUUCCCCGCUUUCAAUCGGCCCGUUCAACU-CGCUUUCAAUCUC ...(((((.(((((.-................)))))----...(..(((((((((((..............)))))))))))..).........-.)))))....... ( -23.37, z-score = -2.33, R) >droYak2.chrX 16748954 100 - 21770863 ---AAAGCUGGGAAA-UUUCAGCAGCACAAUGUUCCC----UCUUUUCGAUUGAAAGCCAUUCAAUUUCCCCGCCUUCAAUUGGCCCAUUCAACU-CGCUUUCAAUUUC ---...((((((...-.))))))..............----.......((((((((((..............(((.......)))..........-.)))))))))).. ( -19.01, z-score = -1.13, R) >droEre2.scaffold_4690 8449014 101 - 18748788 ---AAAGCUGGGAAG-UUUCAGGCCCACAAUGUUCCCUGCUUUUCUCCGAUUGAAAGCC---GAAUUUCCGCGCUUUCAAUCGGCCCGUUCAACU-CGCCUUCAAUCCC ---(((((.(((((.-................))))).)))))...(((((((((((((---(......)).))))))))))))...........-............. ( -29.93, z-score = -2.82, R) >droAna3.scaffold_13248 3710256 107 + 4840945 -ACAAUAAAGGGAAAUUUGAAGGGAAAUUAAUUUCCCUAC-UUCACCCAACAAGUUGAAAUACGAAAAGCUUUCAACCAUGUGGCAAGUUGAAAGGGGCAUUCAACAAA -........(((......(.(((((((....))))))).)-....))).....((((((...(.....)((((((((..........)))))))).....))))))... ( -23.60, z-score = -1.23, R) >dp4.chrXL_group3a 89811 90 - 2690836 -GCGAGACAGGGAAA-UUCCAG--ACAAAAUGUUCCUACUUAUUCGCUGGUUGAAUGCG---ACAUUCAACCCCGAUUGAAAGCAUCGUUCAACU--GC---------- -((((..((((((.(-((....--....))).)))))..(((.(((..(((((((((..---.))))))))).))).)))..)..))))......--..---------- ( -20.60, z-score = -0.88, R) >droPer1.super_39 98131 90 + 745454 -GCGAGACAGGGAAA-UUCCAG--ACAAAAUGUUCCUACUUAUUCGCUGGUUGAAUGCG---ACAUUCAACCCCGAUUGAAAGCAUCGUUCAACU--GC---------- -((((..((((((.(-((....--....))).)))))..(((.(((..(((((((((..---.))))))))).))).)))..)..))))......--..---------- ( -20.60, z-score = -0.88, R) >consensus ___AAAGCUGGGAAA_UUUCAGCCCCACAAUGUUCCC____UCUCUCCGAUUGAAAGCCA__GAAUUUCCCCGCUUUCAAUCGGCCCGUUCAACU_CGCUUUCAAUCUC .........(((((..................))))).........(((.((((((((..............)))))))).)))......................... ( -7.94 = -7.61 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:38 2011