
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,100,270 – 18,100,405 |
| Length | 135 |
| Max. P | 0.745288 |

| Location | 18,100,270 – 18,100,373 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.17 |
| Shannon entropy | 0.55128 |
| G+C content | 0.45264 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.37 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.512075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
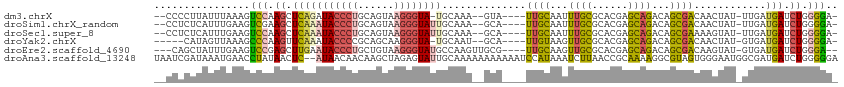
>dm3.chrX 18100270 103 - 22422827 --CCCCUUAUUUAAAGUCCAAGCUCAGAUACCCUGCAGUAAGGGUA-UGCAAA--GUA----UUGCAAUUUGCGCACGAGCAGACAGCGACAACUAU-UUGAUGAUCUGGGGA- --((((.........(((...((((..(((((((......))))))-)(((((--...----......)))))....)))).)))...((((.(...-..).)).)).)))).- ( -30.00, z-score = -1.85, R) >droSim1.chrX_random 202544 104 - 5698898 --CCUCUCAUUUGAAGUCGAAGCUCAAAUACCCUGCAGUAAGGGUAUUGCAAA--GCA----UUGCAAUUUGCGCACGAGCAGACAGCGACAACUAU-UUGAUGAUCUGGGGA- --(((((((((.(((((.(..(((..((((((((......))))))))....)--)).----((((..(((((......)))))..))))).))).)-).)))))...)))).- ( -31.20, z-score = -1.71, R) >droSec1.super_8 412355 104 - 3762037 --CCUCUCAUUUGAAGUCCAAGCUCAAAUACCCUGCAGUAAGGGUAUUGCAAA--GCA----UUGCAAUUUGCGCACGAGCAGACAGCGAAAAGUAU-UUGAUGAUCUGGGGA- --(((((((((.((.......(((..((((((((......))))))))....)--)).----((((..(((((......)))))..))))......)-).)))))...)))).- ( -30.20, z-score = -1.34, R) >droYak2.chrX 16726460 100 - 21770863 -----CAUAGUUAAAGCCCAAGUUCAAAUACCCCGCAGCAAGGGUA-UGCAAU--GCA----UUGUAAGUUGCGCACGAGCAGACAGCGACAACUAU-GUGAUGAUCUGGGGA- -----...........((((.......((((((........)))))-).....--.((----(..((((((((((...........))).))))).)-)..)))...))))..- ( -25.80, z-score = -0.23, R) >droEre2.scaffold_4690 8430504 104 - 18748788 ---CAGCUAUUUGAAGUCCGAGCUUGAAUACCCUGCUGUAAGGGUAUGCCAAGUUGCG----UUGCAAGUUGCGCACGAGCAGACAGCGACAAGUAU-GUGAUGAUCUGGGA-- ---.............((((((((((.(((((((......)))))))..))))))..(----((((...((((......))))...)))))......-.........)))).-- ( -30.80, z-score = -0.38, R) >droAna3.scaffold_13248 3684322 112 + 4840945 UAAUCGAUAAAUGAACCUAUAACUC--AUAACAACAAGCUAGAGUAUUGCAAAAAAAAAAAUCCAUAAAUCUUAACCGCAAAAGGCGUAGUGGGAAUGGCGAUGAUCUGGGGGA ..........((((.........))--)).........(((((.((((((...................(((((..(((.....)))...)))))...)))))).))))).... ( -15.55, z-score = 0.90, R) >consensus __CCCCUUAUUUGAAGUCCAAGCUCAAAUACCCUGCAGUAAGGGUAUUGCAAA__GCA____UUGCAAUUUGCGCACGAGCAGACAGCGACAACUAU_GUGAUGAUCUGGGGA_ ................(((.((.(((((((((((......))))))))..............((((...((((......))))...))))............))).)).))).. (-15.16 = -15.37 + 0.20)
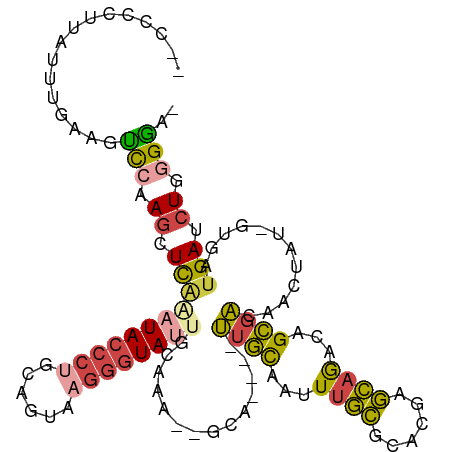
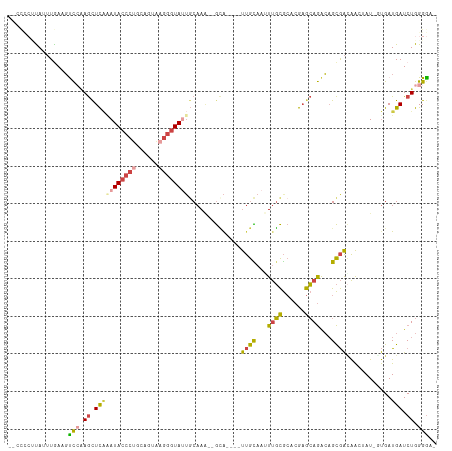
| Location | 18,100,299 – 18,100,405 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 62.72 |
| Shannon entropy | 0.66502 |
| G+C content | 0.42168 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -8.50 |
| Energy contribution | -9.59 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18100299 106 - 22422827 AACAGAGAUAGAGAG-----UAUAAGCUAUGUUAUACCCCCUUAUUUAAAGUCCAAGCUCAGAUACCCUGCAGUAAGGGUA-UGCAA---AGUAUUGCAAUUUGCGCACGAGCAG .....((((((.(.(-----(((((......))))))..).)))))).........((((..(((((((......))))))-)((((---(.........)))))....)))).. ( -25.10, z-score = -1.94, R) >droSim1.chrX_random 202573 107 - 5698898 AACAGAGAUAGAGAG-----UAUAAACUAUGGUAUACCCUCUCAUUUGAAGUCGAAGCUCAAAUACCCUGCAGUAAGGGUAUUGCAA---AGCAUUGCAAUUUGCGCACGAGCAG ..((((...((((.(-----((((........))))).))))..))))........((((.((((((((......))))))))((((---(.........)))))....)))).. ( -31.50, z-score = -3.16, R) >droSec1.super_8 412384 112 - 3762037 AACGAAGAUAGAGAGUACCAUAUAAACUAUGGUAUACCCUCUCAUUUGAAGUCCAAGCUCAAAUACCCUGCAGUAAGGGUAUUGCAA---AGCAUUGCAAUUUGCGCACGAGCAG .((..((((.((((((((((((.....)))))))....)))))))))...))....((((.((((((((......))))))))((((---(.........)))))....)))).. ( -35.10, z-score = -4.39, R) >droYak2.chrX 16726489 96 - 21770863 -------CCAGAGAG-----UAAUAAAUAUGGUAUAACA--U-AGUUAAAGCCCAAGUUCAAAUACCCCGCAGCAAGGGUA-UGCAA---UGCAUUGUAAGUUGCGCACGAGCAG -------.....(.(-----(..(((.((((......))--)-).)))..)).)..((((..((((((........)))))-)((((---(.........)))))....)))).. ( -19.10, z-score = -0.66, R) >droEre2.scaffold_4690 8430532 101 - 18748788 -------UUGGAGAA-----UAAUAACUAUGCUAAACCAGCU-AUUUGAAGUCCGAGCUUGAAUACCCUGCUGUAAGGGUA-UGCCAAGUUGCGUUGCAAGUUGCGCACGAGCAG -------(((((...-----.((((.....(((.....))))-))).....)))))(((((.(((((((......))))))-)(((((.((((...)))).))).)).))))).. ( -29.70, z-score = -1.68, R) >droPer1.super_39 78744 86 + 745454 ---------------------AGCAAUU-UCAAAGUUCGCAGGGUCUGUCAUCGAAUGCUGAAGCUCCU--UAUUAGGGUA-UUUA----GACAUUAUAAUUUGCGCACGAGCAA ---------------------.......-.....(((((..(.((..((.....(((((((((..((((--....))))..-))))----).))))...))..)).).))))).. ( -17.00, z-score = 0.26, R) >dp4.chrXL_group3a 71470 86 - 2690836 ---------------------AGCAAUU-UCAAAGUUCGCAGGGUCUGUCAUCGAAUGCUGAAGCUCCU--UAUUAGGGUA-UUUA----GACAUUAUAAUUUGCGCACGAGCAA ---------------------.......-.....(((((..(.((..((.....(((((((((..((((--....))))..-))))----).))))...))..)).).))))).. ( -17.00, z-score = 0.26, R) >consensus _______AUAGAGAG_____UAUAAACUAUGGUAUACCCCCU_AUUUGAAGUCCAAGCUCAAAUACCCUGCAGUAAGGGUA_UGCAA___AGCAUUGCAAUUUGCGCACGAGCAG ........................................................((((...((((((......))))))..........(((........)))....)))).. ( -8.50 = -9.59 + 1.09)

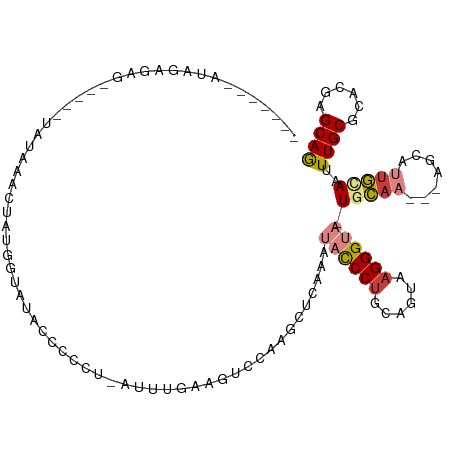
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:35 2011