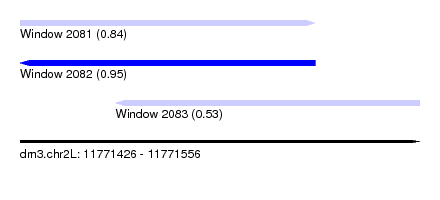
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,771,426 – 11,771,556 |
| Length | 130 |
| Max. P | 0.953350 |

| Location | 11,771,426 – 11,771,522 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.50592 |
| G+C content | 0.43045 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -10.00 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11771426 96 + 23011544 -----UAUUAUAUUAU-UUUUAAGUGCCAACCAUGUGCGCACUUAACAACACCGGAAGCAACUGCCAAAGA-GCUGCUUCAACUGCUUC--UUCAGCUGCUUUUC------------ -----...........-..((((((((((......)).)))))))).......(((((((.(((...((((-((..........)).))--))))).))))))).------------ ( -21.80, z-score = -1.54, R) >droSim1.chr2L 11581478 96 + 22036055 -----UAUUAUAUUAU-UUUUAAGUGCCAACCAUGUGCGCACUUAACAACACCGGAAGCAACUGCCAAAGC-GCUGCUUCCACUGCUUC--UUCAGCUGCUUUUC------------ -----...........-..((((((((((......)).)))))))).......(((((((..(((....))-).)))))))...(((..--...)))........------------ ( -23.50, z-score = -2.20, R) >droSec1.super_3 7160767 96 + 7220098 -----UAUUAUAUUAU-UUUUAAGUGCCAACCAUGUGCGCACUUAACAACACCGGAAGCAACUGCCAAAGC-GCUGCUUCCACUGCUUC--UUCAGCUGCUUUUC------------ -----...........-..((((((((((......)).)))))))).......(((((((..(((....))-).)))))))...(((..--...)))........------------ ( -23.50, z-score = -2.20, R) >droYak2.chr2L 8191663 96 + 22324452 -----UAUUAUAUUAU-UUUUAAGUGCCAACCAUGUGUGCACUUAACAACACCGGAAGCAACUGCCAAAGC-GCUGCUUCAACUGCUUC--UUCAGCUGCUUUUC------------ -----...........-..((((((((((......)).)))))))).......(((((((.(((...((((-(.(......).))))).--..))).))))))).------------ ( -22.30, z-score = -1.64, R) >droAna3.scaffold_12916 9478094 111 - 16180835 -----UAUUAUAUUAU-UUUUAAGUGCCAACCAUGUGCGCACUUAACAACACCGGAAGCAGCUUCUUGGGCAUCUUGGCCUUUUACUUCGCUCUACCCCCUACUCUACCCCCUUCUC -----...........-..((((((((((......)).)))))))).......((.((.(((.....((((......))))........))))).)).................... ( -19.62, z-score = -1.31, R) >dp4.chr4_group3 2643656 76 + 11692001 -----UAUUAUAUUAU-UUUUAAGUGCCAACCAUGUGCGCACUUAACAACACCGGAAGCAACUGCCAAAGCAGCUGCCAUAC----------------------------------- -----...........-..((((((((((......)).))))))))...........(((.((((....)))).))).....----------------------------------- ( -18.00, z-score = -2.10, R) >droPer1.super_1 4118229 76 + 10282868 -----UAUUAUAUUAU-UUUUAAGUGCCAACCAUGUGCGCACUUAACAACACCGGAAGCAACUGCCAAAGCAGCUGCCAUAC----------------------------------- -----...........-..((((((((((......)).))))))))...........(((.((((....)))).))).....----------------------------------- ( -18.00, z-score = -2.10, R) >droWil1.scaffold_180772 3649764 83 + 8906247 UUUUUUUUUAUUUCUCAUUUUAAGUGCCAGCCAUGUGCGCACUUAACAAUACCGGAAGGAGCC----AGGGAACUAGGAUGGUUGGC------------------------------ .........................(((((((((.................((....))..((----((....)).)))))))))))------------------------------ ( -22.20, z-score = -1.31, R) >consensus _____UAUUAUAUUAU_UUUUAAGUGCCAACCAUGUGCGCACUUAACAACACCGGAAGCAACUGCCAAAGC_GCUGCUUCAACUGCUUC__UUCAGCUGCUUUUC____________ ...................((((((((((......)).))))))))......(....)........................................................... (-10.00 = -10.00 + 0.00)

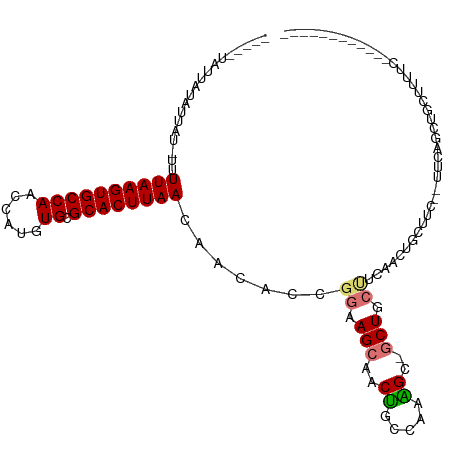
| Location | 11,771,426 – 11,771,522 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.50592 |
| G+C content | 0.43045 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.59 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11771426 96 - 23011544 ------------GAAAAGCAGCUGAA--GAAGCAGUUGAAGCAGC-UCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCGCACAUGGUUGGCACUUAAAA-AUAAUAUAAUA----- ------------...(((((((((..--((((.(((((...))))-)))))..)))))))))...((((((((((((((.((......)))))))))...-)))))))....----- ( -32.40, z-score = -2.65, R) >droSim1.chr2L 11581478 96 - 22036055 ------------GAAAAGCAGCUGAA--GAAGCAGUGGAAGCAGC-GCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCGCACAUGGUUGGCACUUAAAA-AUAAUAUAAUA----- ------------...(((((((((..--(((((..((....))..-)))))..)))))))))...((((((((((((((.((......)))))))))...-)))))))....----- ( -33.30, z-score = -2.65, R) >droSec1.super_3 7160767 96 - 7220098 ------------GAAAAGCAGCUGAA--GAAGCAGUGGAAGCAGC-GCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCGCACAUGGUUGGCACUUAAAA-AUAAUAUAAUA----- ------------...(((((((((..--(((((..((....))..-)))))..)))))))))...((((((((((((((.((......)))))))))...-)))))))....----- ( -33.30, z-score = -2.65, R) >droYak2.chr2L 8191663 96 - 22324452 ------------GAAAAGCAGCUGAA--GAAGCAGUUGAAGCAGC-GCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCACACAUGGUUGGCACUUAAAA-AUAAUAUAAUA----- ------------...(((((((((..--(((((.((((...))))-)))))..)))))))))...((((((((((((((.((......)))))))))...-)))))))....----- ( -33.70, z-score = -3.14, R) >droAna3.scaffold_12916 9478094 111 + 16180835 GAGAAGGGGGUAGAGUAGGGGGUAGAGCGAAGUAAAAGGCCAAGAUGCCCAAGAAGCUGCUUCCGGUGUUGUUAAGUGCGCACAUGGUUGGCACUUAAAA-AUAAUAUAAUA----- .....((((((((..(...(((((...(...((.....))...).)))))...)..)))))))).((((((((((((((.((......)))))))))...-)))))))....----- ( -32.30, z-score = -2.37, R) >dp4.chr4_group3 2643656 76 - 11692001 -----------------------------------GUAUGGCAGCUGCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCGCACAUGGUUGGCACUUAAAA-AUAAUAUAAUA----- -----------------------------------....(((((((((....)))))))))....((((((((((((((.((......)))))))))...-)))))))....----- ( -26.20, z-score = -2.36, R) >droPer1.super_1 4118229 76 - 10282868 -----------------------------------GUAUGGCAGCUGCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCGCACAUGGUUGGCACUUAAAA-AUAAUAUAAUA----- -----------------------------------....(((((((((....)))))))))....((((((((((((((.((......)))))))))...-)))))))....----- ( -26.20, z-score = -2.36, R) >droWil1.scaffold_180772 3649764 83 - 8906247 ------------------------------GCCAACCAUCCUAGUUCCCU----GGCUCCUUCCGGUAUUGUUAAGUGCGCACAUGGCUGGCACUUAAAAUGAGAAAUAAAAAAAAA ------------------------------((((.((((.((((....))----)).........(((((....)))))....)))).))))......................... ( -15.50, z-score = 0.06, R) >consensus ____________GAAAAGCAGCUGAA__GAAGCAGUUGAAGCAGC_GCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCGCACAUGGUUGGCACUUAAAA_AUAAUAUAAUA_____ .......................................((((((..........))))))....((((((((((((((.((......)))))))))....)))))))......... (-13.09 = -13.59 + 0.50)
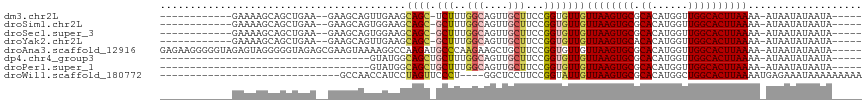
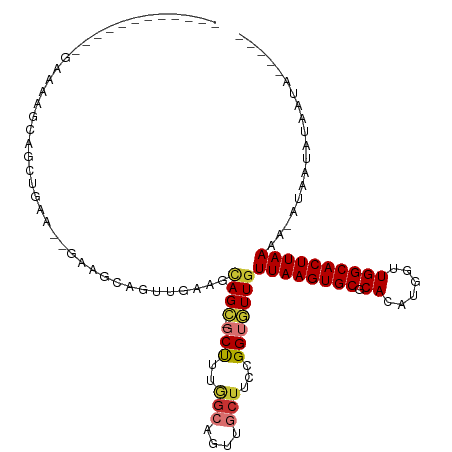
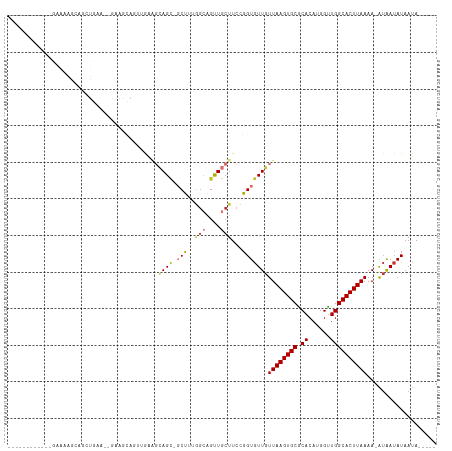
| Location | 11,771,457 – 11,771,556 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Shannon entropy | 0.37035 |
| G+C content | 0.50734 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -19.07 |
| Energy contribution | -22.28 |
| Covariance contribution | 3.21 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11771457 99 - 23011544 CCAAAUUGCUUGAGAAGCAUGCU-----AGCAAAGCACGGAAAAGCAGCUGAAGAAGCAGUUGAAGCAGCUCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCG ................((((..(-----(((((.((.((((..((((((((..((((.(((((...)))))))))..)))))))))))))).)))))).)))). ( -38.40, z-score = -2.46, R) >droSim1.chr2L 11581509 99 - 22036055 CCAAAUUGCUUGAGAAGCAUGCU-----AGGAAAGCACGGAAAAGCAGCUGAAGAAGCAGUGGAAGCAGCGCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCG .......((((((..((((((((-----.....))).((((..((((((((..(((((..((....))..)))))..))))))))))))))))).))))))... ( -37.30, z-score = -2.20, R) >droSec1.super_3 7160798 99 - 7220098 CCAAAUUGCUUGAGAAGCAUGCU-----AGCAAAGCACGGAAAAGCAGCUGAAGAAGCAGUGGAAGCAGCGCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCG ................((((..(-----(((((.((.((((..((((((((..(((((..((....))..)))))..)))))))))))))).)))))).)))). ( -39.30, z-score = -2.44, R) >droYak2.chr2L 8191694 104 - 22324452 CCAAAUUGCUUGAGAAGCAUGCUAGCAAAGCAAAGCACGGAAAAGCAGCUGAAGAAGCAGUUGAAGCAGCGCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCA ......((((.....)))).....(((.(((((.((.((((..((((((((..(((((.((((...)))))))))..)))))))))))))).)))))...))). ( -38.70, z-score = -1.77, R) >droWil1.scaffold_180772 3649801 82 - 8906247 CCAAAUUGCUUUGAAGCCGCUC--------AGAUGCCC-------CACCCGGCCAACCAUCC----UAGUUCCCUGGC---UCCUUCCGGUAUUGUUAAGUGCG .......(((....)))((((.--------.((((((.-------.....(((((.......----........))))---)......))))))....)))).. ( -14.66, z-score = 0.51, R) >consensus CCAAAUUGCUUGAGAAGCAUGCU_____AGCAAAGCACGGAAAAGCAGCUGAAGAAGCAGUGGAAGCAGCGCUUUGGCAGUUGCUUCCGGUGUUGUUAAGUGCG .......((((((..((((((((.....)))......((((..((((((((..(((((.((.......)))))))..))))))))))))))))).))))))... (-19.07 = -22.28 + 3.21)

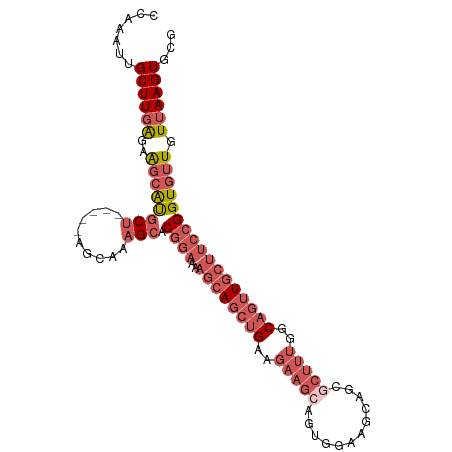
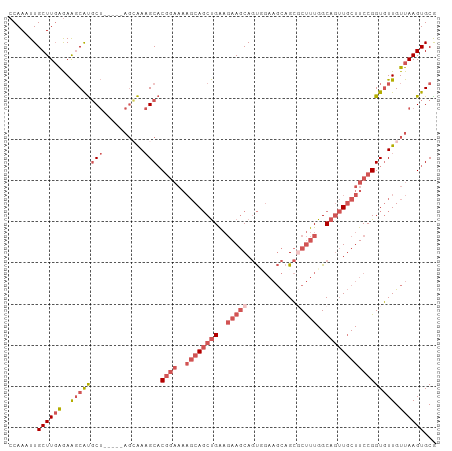
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:37 2011