| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,075,419 – 18,075,545 |
| Length | 126 |
| Max. P | 0.976762 |

| Location | 18,075,419 – 18,075,519 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.46 |
| Shannon entropy | 0.45057 |
| G+C content | 0.46994 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -19.30 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
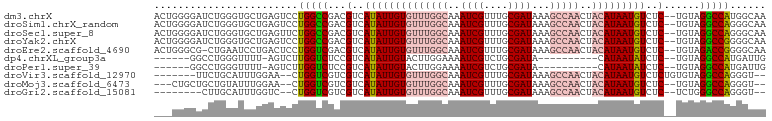
>dm3.chrX 18075419 100 - 22422827 ACUGGGGAUCUGGGUGCUGAGUCCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC--UGUAGGCCAUGGCAA (((.((.((....)).)).)))(((((((.(((..((((((((((((((..((((....))))...)))))..)))))))))...--))).))))).))... ( -34.40, z-score = -1.88, R) >droSim1.chrX_random 177115 100 - 5698898 ACUGGGGAUCUGGGUGCUGAGUCCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC--UGUAGGCCAGGGCAA .(..(....)..).......(((((((((.(((..((((((((((((((..((((....))))...)))))..)))))))))...--))).))))))))).. ( -40.30, z-score = -3.45, R) >droSec1.super_8 386936 100 - 3762037 ACUGGGGAUCUGGGUGCUGAGUUCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC--UGUAGGCCAGGGCAA .(..(....)..).......(((((((((.(((..((((((((((((((..((((....))))...)))))..)))))))))...--))).))))))))).. ( -37.50, z-score = -2.90, R) >droYak2.chrX 16702511 100 - 21770863 ACUGGGGAUCUGGGUGCUGAGUCCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC--UGUAGGCCGGGGCAA .(..(....)..).......(((((((((.(((..((((((((((((((..((((....))))...)))))..)))))))))...--))).))))))))).. ( -39.60, z-score = -3.10, R) >droEre2.scaffold_4690 8406490 99 - 18748788 ACUGGGCG-CUGAAUCCUGACUCCUGGUCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC--UGUAGACCGGGGCAA ...(((..-......)))...((((((((.(((..((((((((((((((..((((....))))...)))))..)))))))))...--))).))))))))... ( -32.30, z-score = -2.30, R) >dp4.chrXL_group3a 40702 83 - 2690836 ------GGCCUGGGUUUU-AGUCUUGGUCUCCGUCAUAUUGUACUUGGAAAAUCGUCUGCGAUA----------CAUAAUAUCUC--UGUAGGCCAUGAUUG ------...........(-((((.(((((((.(..(((((((...(....)((((....)))).----------.)))))))..)--.).)))))).))))) ( -16.10, z-score = 0.34, R) >droPer1.super_39 47149 83 + 745454 ------GGCCUGGGUUUU-AGUCUUGGUCUCCGUCAUAUUGUACUUGGAAAAUCGUCUGCGAUA----------CAUAAUAUCUC--UGUAGGCCAUGAUUG ------...........(-((((.(((((((.(..(((((((...(....)((((....)))).----------.)))))))..)--.).)))))).))))) ( -16.10, z-score = 0.34, R) >droVir3.scaffold_12970 9131326 91 + 11907090 -------UUCUGCAUUUGGAA--CUGGUCGUCGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCUGUGUAGGCCAGGGU-- -------((((......))))--((((((..((..((((((((((((((..((((....))))...)))))..)))))))))...))....))))))...-- ( -27.00, z-score = -2.12, R) >droMoj3.scaffold_6473 13401079 93 + 16943266 ---CUGCUGCUGUAUUUGGAA--CUGGUCGUCGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC--UGUAGGCCAGGGU-- ---.(((....))).......--((((((...(..((((((((((((((..((((....))))...)))))..)))))))))..)--....))))))...-- ( -27.20, z-score = -1.71, R) >droGri2.scaffold_15081 1022846 88 - 4274704 --------CUUGCAUUUGGUC--CUGGUCGUCGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC--UCUGGGCCAGGGU-- --------...........((--((((((...(..((((((((((((((..((((....))))...)))))..)))))))))..)--....)))))))).-- ( -29.30, z-score = -3.36, R) >consensus ___GGGGAUCUGGGUUCUGAGUCCUGGUCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUC__UGUAGGCCAGGGCAA ........................(((((...(..((((((((((((((..((((....))))...)))))..)))))))))..)......)))))...... (-19.30 = -20.05 + 0.75)

| Location | 18,075,433 – 18,075,545 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Shannon entropy | 0.28361 |
| G+C content | 0.46479 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -15.77 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18075433 112 + 22422827 AGAGACAUUAUGUAGUUGGCUUUAUCGCAAACGAUUUGCCAAACACAAUAUGACGUCGGCCAGGACUCAGCACCCAGAUCCCCAGUUUCCCAGCUUCCCAGUUACCCAGCAU .(((((....(((..(((((...((((....))))..)))))..)))...(((.(((......))))))...............)))))...(((............))).. ( -22.70, z-score = -1.15, R) >droSim1.chrX_random 177129 87 + 5698898 AGAGACAUUAUGUAGUUGGCUUUAUCGCAAACGAUUUGCCAAACACAAUAUGACGUCGGCCAGGACUCAGCACCCAGAUCCCCAGUU------------------------- ...(((.((((((.((((((...((((....))))..))))...)).)))))).)))((...(((.((........)))))))....------------------------- ( -20.10, z-score = -1.48, R) >droSec1.super_8 386950 87 + 3762037 AGAGACAUUAUGUAGUUGGCUUUAUCGCAAACGAUUUGCCAAACACAAUAUGACGUCGGCCAGAACUCAGCACCCAGAUCCCCAGUU------------------------- ...(((.((((((.((((((...((((....))))..))))...)).)))))).)))((...((.((........)).)).))....------------------------- ( -17.20, z-score = -0.98, R) >droYak2.chrX 16702525 87 + 21770863 AGAGACAUUAUGUAGUUGGCUUUAUCGCAAACGAUUUGCCAAACACAAUAUGACGUCGGCCAGGACUCAGCACCCAGAUCCCCAGUU------------------------- ...(((.((((((.((((((...((((....))))..))))...)).)))))).)))((...(((.((........)))))))....------------------------- ( -20.10, z-score = -1.48, R) >droEre2.scaffold_4690 8406504 86 + 18748788 AGAGACAUUAUGUAGUUGGCUUUAUCGCAAACGAUUUGCCAAACACAAUAUGACGUCGACCAGGAGUCAGGAUUCAG-CGCCCAGUU------------------------- .(((.(....(((..(((((...((((....))))..)))))..)))...((((..(.....)..)))).).)))..-.........------------------------- ( -19.00, z-score = -0.84, R) >droGri2.scaffold_15081 1022858 86 + 4274704 AGAGACAUUAUGUAGUUGGCUUUAUCGCAAACGAUUUGCCAAACACAAUAUGACGACGACCAGGACCAA--AUGCAAGCAGCAAGCAG------------------------ .....(((..(((..(((((...((((....))))..)))))..)))..))).................--.(((.....))).....------------------------ ( -16.20, z-score = -0.83, R) >consensus AGAGACAUUAUGUAGUUGGCUUUAUCGCAAACGAUUUGCCAAACACAAUAUGACGUCGGCCAGGACUCAGCACCCAGAUCCCCAGUU_________________________ ...(((.((((((.((((((...((((....))))..))))...)).)))))).)))....................................................... (-15.77 = -15.93 + 0.17)

| Location | 18,075,433 – 18,075,545 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Shannon entropy | 0.28361 |
| G+C content | 0.46479 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18075433 112 - 22422827 AUGCUGGGUAACUGGGAAGCUGGGAAACUGGGGAUCUGGGUGCUGAGUCCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCU ..((..((..(((.(((..(..(....)..)...))).))).......))..)).(((((.....((((.(((((..((((....))))...))))).))))..)))))... ( -35.40, z-score = -1.48, R) >droSim1.chrX_random 177129 87 - 5698898 -------------------------AACUGGGGAUCUGGGUGCUGAGUCCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCU -------------------------..(..((..((........))..))..)..(((((.....((((.(((((..((((....))))...))))).))))..)))))... ( -24.60, z-score = -1.09, R) >droSec1.super_8 386950 87 - 3762037 -------------------------AACUGGGGAUCUGGGUGCUGAGUUCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCU -------------------------((((.((.((....)).)).))))..(((....)))((((((((((((((..((((....))))...)))))..))))))))).... ( -23.20, z-score = -0.83, R) >droYak2.chrX 16702525 87 - 21770863 -------------------------AACUGGGGAUCUGGGUGCUGAGUCCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCU -------------------------..(..((..((........))..))..)..(((((.....((((.(((((..((((....))))...))))).))))..)))))... ( -24.60, z-score = -1.09, R) >droEre2.scaffold_4690 8406504 86 - 18748788 -------------------------AACUGGGCG-CUGAAUCCUGACUCCUGGUCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCU -------------------------.....((((-.(((..((........))))).))))((((((((((((((..((((....))))...)))))..))))))))).... ( -23.60, z-score = -2.11, R) >droGri2.scaffold_15081 1022858 86 - 4274704 ------------------------CUGCUUGCUGCUUGCAU--UUGGUCCUGGUCGUCGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCU ------------------------(((..(((.....))).--.)))...........(..((((((((((((((..((((....))))...)))))..)))))))))..). ( -19.80, z-score = -0.89, R) >consensus _________________________AACUGGGGAUCUGGGUGCUGAGUCCUGGCCGACGUCAUAUUGUGUUUGGCAAAUCGUUUGCGAUAAAGCCAACUACAUAAUGUCUCU ...........................(..(((.((........)).)))..).....(..((((((((((((((..((((....))))...)))))..)))))))))..). (-20.25 = -20.75 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:26 2011