| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,069,816 – 18,069,913 |
| Length | 97 |
| Max. P | 0.993949 |

| Location | 18,069,816 – 18,069,913 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.28 |
| Shannon entropy | 0.53123 |
| G+C content | 0.39460 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -10.30 |
| Energy contribution | -10.76 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
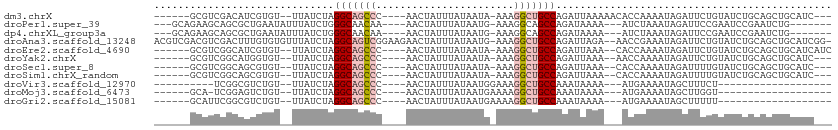
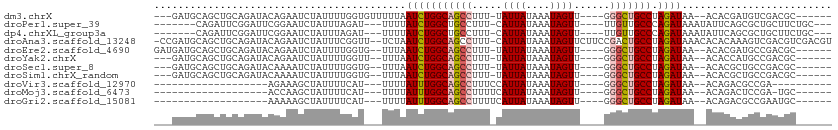
>dm3.chrX 18069816 97 + 22422827 ------GCGUCGACAUCGUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUA-AAAGGCUGCCAGAUUAAAAACACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUC--- ------(((.(......((((--((((((.(((((((.----..............-...)))))))))))...)))))).....(((((.....)))))..).)))...--- ( -25.37, z-score = -3.02, R) >droPer1.super_39 41890 95 - 745454 ---GCAGAAGCAGCGCUGAAUAUUUAUCUGGGCAACAA----AACUAUUUAUAAUG-AAAGGCAGCCAGAUAAAA---AUCUAAAUAGAUUCCGAAUCCGAAUCUG------- ---..((((((...))).....(((((((((((.....----..............-....))..))))))))).---.)))...(((((((.......)))))))------- ( -15.30, z-score = -0.04, R) >dp4.chrXL_group3a 35579 95 + 2690836 ---GCAGAAGCAGCGCUGAAUAUUUAUCUGGGCAACAA----AACUAUUUAUAAUG-AAAGGCAGCCAGAUAAAA---AUCUAAAUAGAUUCCGAAUCCGAAUCUG------- ---..((((((...))).....(((((((((((.....----..............-....))..))))))))).---.)))...(((((((.......)))))))------- ( -15.30, z-score = -0.04, R) >droAna3.scaffold_13248 3640577 109 - 4840945 ACGUCGACGUCGACUUUGUGUGUUUAUCUAGGCAGUCGGAAGAACUAUUUAUAAUG-AAAGGCUGCCAGAUUAGA--AACCGAAAUAGAUUCUGUAUCUGCAGCUGCAUCGG- ..((((....))))........(((((((.(((((((...................-...)))))))))).))))--..((((..(((...((((....)))))))..))))- ( -26.15, z-score = -0.15, R) >droEre2.scaffold_4690 8398104 98 + 18748788 ------GCGUCGGCAUCGUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUA-AAAGGCUGCCAGAUUAAA--CACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUCAUC ------..(.((((...((((--((((((.(((((((.----..............-...)))))))))).))))--))).....(((((.....)))))..)))))...... ( -30.17, z-score = -3.98, R) >droYak2.chrX 16695801 95 + 21770863 ------GCGUCGGCAUGGUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUA-AAAGGCUGCCAGAUUAAA--AACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUC--- ------..(.((((.((((.(--((((((.(((((((.----..............-...)))))))))).))))--.))))...(((((.....)))))..)))))...--- ( -29.07, z-score = -3.08, R) >droSec1.super_8 381349 95 + 3762037 ------GCGUCGGCAGCGUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUA-AAAGGCUGCCAGAUUAAA--CACCAAAAUAGAUUUUGUAUCUGCAGCUGCAUC--- ------......(((((((((--((((((.(((((((.----..............-...)))))))))).))))--))).....(((((.....)))))..)))))...--- ( -34.37, z-score = -4.88, R) >droSim1.chrX_random 171556 95 + 5698898 ------GCGUCGGCAGCGUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUA-AAAGGCUGCCAGAUUAAA--CACCAAAAUAGAUUUUGUAUCUGCAGCUGCAUC--- ------......(((((((((--((((((.(((((((.----..............-...)))))))))).))))--))).....(((((.....)))))..)))))...--- ( -34.37, z-score = -4.88, R) >droVir3.scaffold_12970 2768792 75 + 11907090 ----------UCGGCGUCUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUGGAAAGGCUGCCAAAUAAAA---AUGAAAAUAGCUUUCU------------------- ----------..(((.((..(--((((...(((((((.----..((((.....))))...)))))))..))))).---..)).....)))....------------------- ( -19.80, z-score = -2.45, R) >droMoj3.scaffold_6473 3533361 78 + 16943266 ------GCA-UCGGAGUCUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUGAAAAGGCUGCCAAAUAAAA---AUGAAAAUAGCUUGGU------------------- ------...-.(.(((.((((--((.((..(((((((.----..................)))))))........---..))))))))))).).------------------- ( -16.91, z-score = -0.96, R) >droGri2.scaffold_15081 3245769 79 - 4274704 ------GCAUUCGGCGUCUGU--UUAUCUAGGCAGCCC----AACUAUUUAUAAUGAAAAGGCUGCCAAAUAAAA---AUGAAAAUAGCUUUUU------------------- ------......(((.((..(--((((...(((((((.----..................)))))))..))))).---..)).....)))....------------------- ( -16.71, z-score = -1.01, R) >consensus ______GCGUCGGCAGUGUGU__UUAUCUAGGCAGCCC____AACUAUUUAUAAUG_AAAGGCUGCCAGAUUAAA___ACCAAAAUAGAUUCUGUAUCUGCAGCUGCAUC___ ..............................(((((((.......................))))))).............................................. (-10.30 = -10.76 + 0.46)
| Location | 18,069,816 – 18,069,913 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.28 |
| Shannon entropy | 0.53123 |
| G+C content | 0.39460 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.59 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.993949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
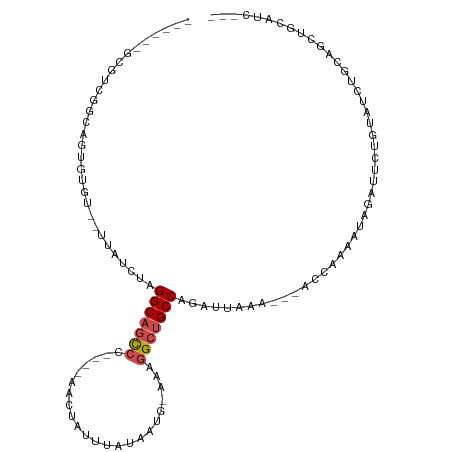
>dm3.chrX 18069816 97 - 22422827 ---GAUGCAGCUGCAGAUACAGAAUCUAUUUUGGUGUUUUUAAUCUGGCAGCCUUU-UAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACACGAUGUCGACGC------ ---..(((....)))(((((((((....)))))((((((...((((((((((((..-.((((....)))).----)))))))).))))))--))))..)))).....------ ( -30.80, z-score = -3.31, R) >droPer1.super_39 41890 95 + 745454 -------CAGAUUCGGAUUCGGAAUCUAUUUAGAU---UUUUAUCUGGCUGCCUUU-CAUUAUAAAUAGUU----UUGUUGCCCAGAUAAAUAUUCAGCGCUGCUUCUGC--- -------((((..(((.....((((((....))))---))((((((((..((....-((..((.....)).----.))..))))))))))..........)))..)))).--- ( -18.70, z-score = -0.26, R) >dp4.chrXL_group3a 35579 95 - 2690836 -------CAGAUUCGGAUUCGGAAUCUAUUUAGAU---UUUUAUCUGGCUGCCUUU-CAUUAUAAAUAGUU----UUGUUGCCCAGAUAAAUAUUCAGCGCUGCUUCUGC--- -------((((..(((.....((((((....))))---))((((((((..((....-((..((.....)).----.))..))))))))))..........)))..)))).--- ( -18.70, z-score = -0.26, R) >droAna3.scaffold_13248 3640577 109 + 4840945 -CCGAUGCAGCUGCAGAUACAGAAUCUAUUUCGGUU--UCUAAUCUGGCAGCCUUU-CAUUAUAAAUAGUUCUUCCGACUGCCUAGAUAAACACACAAAGUCGACGUCGACGU -.(((((..(((((((((...(((((......))))--)...)))).)))))....-..................(((((..................))))).))))).... ( -23.67, z-score = -0.90, R) >droEre2.scaffold_4690 8398104 98 - 18748788 GAUGAUGCAGCUGCAGAUACAGAAUCUAUUUUGGUG--UUUAAUCUGGCAGCCUUU-UAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACACGAUGCCGACGC------ ......((..(.(((....(((((....)))))(((--((((.(((((((((((..-.((((....)))).----)))))))).))))))--))))..))).)..))------ ( -31.80, z-score = -3.30, R) >droYak2.chrX 16695801 95 - 21770863 ---GAUGCAGCUGCAGAUACAGAAUCUAUUUUGGUU--UUUAAUCUGGCAGCCUUU-UAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACACCAUGCCGACGC------ ---...((..(.((((((.....))))....((((.--((((.(((((((((((..-.((((....)))).----)))))))).))))))--).)))).)).)..))------ ( -28.40, z-score = -2.68, R) >droSec1.super_8 381349 95 - 3762037 ---GAUGCAGCUGCAGAUACAAAAUCUAUUUUGGUG--UUUAAUCUGGCAGCCUUU-UAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACACGCUGCCGACGC------ ---...(((((........(((((....)))))(((--((((.(((((((((((..-.((((....)))).----)))))))).))))))--)))))))))......------ ( -36.40, z-score = -4.88, R) >droSim1.chrX_random 171556 95 - 5698898 ---GAUGCAGCUGCAGAUACAAAAUCUAUUUUGGUG--UUUAAUCUGGCAGCCUUU-UAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACACGCUGCCGACGC------ ---...(((((........(((((....)))))(((--((((.(((((((((((..-.((((....)))).----)))))))).))))))--)))))))))......------ ( -36.40, z-score = -4.88, R) >droVir3.scaffold_12970 2768792 75 - 11907090 -------------------AGAAAGCUAUUUUCAU---UUUUAUUUGGCAGCCUUUCCAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACAGACGCCGA---------- -------------------.(((((...)))))..---.(((((((((((((((..(.((.....)).)..----)))))))).))))))--)..........---------- ( -19.20, z-score = -2.16, R) >droMoj3.scaffold_6473 3533361 78 - 16943266 -------------------ACCAAGCUAUUUUCAU---UUUUAUUUGGCAGCCUUUUCAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACAGACUCCGA-UGC------ -------------------................---.(((((((((((((((....((((....)))).----)))))))).))))))--)..........-...------ ( -18.30, z-score = -2.03, R) >droGri2.scaffold_15081 3245769 79 + 4274704 -------------------AAAAAGCUAUUUUCAU---UUUUAUUUGGCAGCCUUUUCAUUAUAAAUAGUU----GGGCUGCCUAGAUAA--ACAGACGCCGAAUGC------ -------------------................---.(((((((((((((((....((((....)))).----)))))))).))))))--)..............------ ( -18.30, z-score = -1.53, R) >consensus ___GAUGCAGCUGCAGAUACAGAAUCUAUUUUGGU___UUUAAUCUGGCAGCCUUU_CAUUAUAAAUAGUU____GGGCUGCCUAGAUAA__ACACACUGCCGACGC______ ..........................................(((((((((((.....((((....))))......))))))).))))......................... (-13.48 = -13.59 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:24 2011