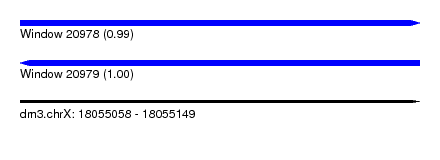
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,055,058 – 18,055,149 |
| Length | 91 |
| Max. P | 0.998565 |

| Location | 18,055,058 – 18,055,149 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.56699 |
| G+C content | 0.33848 |
| Mean single sequence MFE | -17.76 |
| Consensus MFE | -9.97 |
| Energy contribution | -10.29 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
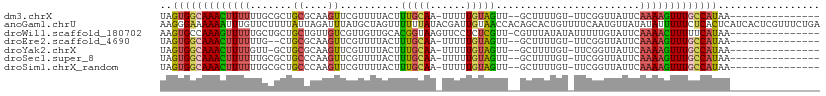
>dm3.chrX 18055058 91 + 22422827 ---------------UUAUGGCAAACUUUUGAAUAACCGAA-ACAAAAGC--AACUACAAAAA-UUGCAAAGUAAAACGAACUUGCGCAGCGCAAAAAAGUUUGCCACUA ---------------...((((((((((((...........-......((--((.........-))))..............(((((...)))))))))))))))))... ( -21.40, z-score = -2.79, R) >anoGam1.chrU 35792705 110 - 59568033 UCAGAAACGAGUGAUGAGUGAGAAAAAUAUAUAACAUUGAAAACAGUGCUGUGGUUACAAUCGUAUAAAAAACUAGCAUAAAUCUAAUAAAAGAACAAAUUUUUUCCCUU ...............(((.(((((((...................(((((((...(((....)))......)).)))))...(((......))).....))))))).))) ( -11.70, z-score = 0.57, R) >droWil1.scaffold_180702 1249484 94 + 4511350 ---------------UUAUGAAAAAGUUUUGAAUACAAAAAUAUAUAAACG-AACGAGAGGAACUUACCGUGCAACAACGACAACAGCAGCAGCAAAAACUUUGGCACUU ---------------...((..((((((((.....................-..(....)........(((......)))......((....)).))))))))..))... ( -10.60, z-score = -0.04, R) >droEre2.scaffold_4690 8383893 89 + 18748788 ---------------UUAUGGCAAACUUUUGAAUAACCGAA-ACAAAAGC--AACUACAAAAA-UUGCAAAGUAAAACGAACUUGCGCAG--CAAAAAAGUUUGCCACUA ---------------...((((((((((((...........-......((--((.........-))))..............((((...)--)))))))))))))))... ( -19.00, z-score = -2.48, R) >droYak2.chrX 16680917 90 + 21770863 ---------------UUAUGGCAAACUUUUGAAUAACCGAA-ACAAAAGC--AACUACAAAAA-UUGCAAAGUAAAACGAACUUGCGCAGC-AACAAAAGUUUGCCACUA ---------------...(((((((((((((..........-......((--((.........-))))...((((.......)))).....-..)))))))))))))... ( -22.00, z-score = -3.69, R) >droSec1.super_8 367146 91 + 3762037 ---------------UUAUGGCAAACUUUUGAAUAACCGAA-ACAAAAGC--AACUACAAAAA-UUGCAAAGUAAAACGAACUUGGGCAGCGCAAAAAAGUUUGCCACUA ---------------...((((((((((((......((...-......((--((.........-)))).((((.......))))))((...))..))))))))))))... ( -19.80, z-score = -2.13, R) >droSim1.chrX_random 4644939 91 + 5698898 ---------------UUAUGGCAAACUUUUGAAUAACCGAA-ACAAAAGC--AACUACAAAAA-UUGCAAAGUAAAACGAACUUGGGCAGCGCAAAAAAGUUUGCCACUA ---------------...((((((((((((......((...-......((--((.........-)))).((((.......))))))((...))..))))))))))))... ( -19.80, z-score = -2.13, R) >consensus _______________UUAUGGCAAACUUUUGAAUAACCGAA_ACAAAAGC__AACUACAAAAA_UUGCAAAGUAAAACGAACUUGCGCAGCGCAAAAAAGUUUGCCACUA ..................((((((((((((........(....).....................(((.((((.......))))..)))......))))))))))))... ( -9.97 = -10.29 + 0.31)
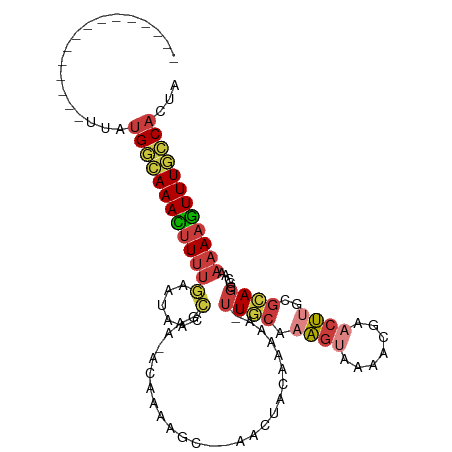
| Location | 18,055,058 – 18,055,149 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.56699 |
| G+C content | 0.33848 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -13.41 |
| Energy contribution | -13.36 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.998565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
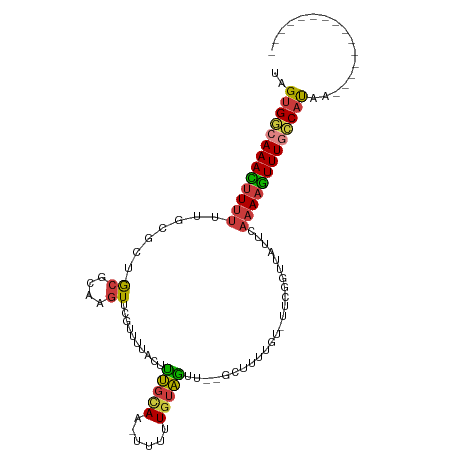
>dm3.chrX 18055058 91 - 22422827 UAGUGGCAAACUUUUUUGCGCUGCGCAAGUUCGUUUUACUUUGCAA-UUUUUGUAGUU--GCUUUUGU-UUCGGUUAUUCAAAAGUUUGCCAUAA--------------- ..((((((((((((((((((...)))))..(((....((...((((-((.....))))--))....))-..)))......)))))))))))))..--------------- ( -26.90, z-score = -3.35, R) >anoGam1.chrU 35792705 110 + 59568033 AAGGGAAAAAAUUUGUUCUUUUAUUAGAUUUAUGCUAGUUUUUUAUACGAUUGUAACCACAGCACUGUUUUCAAUGUUAUAUAUUUUUCUCACUCAUCACUCGUUUCUGA ..((((((((..((((......(((((.......))))).......))))..........((((.((....)).)))).....))))))))................... ( -12.22, z-score = 0.68, R) >droWil1.scaffold_180702 1249484 94 - 4511350 AAGUGCCAAAGUUUUUGCUGCUGCUGUUGUCGUUGUUGCACGGUAAGUUCCUCUCGUU-CGUUUAUAUAUUUUUGUAUUCAAAACUUUUUCAUAA--------------- ..(((..(((((((((((((.(((.............)))))))).............-.(..((((......))))..)))))))))..)))..--------------- ( -14.22, z-score = -0.57, R) >droEre2.scaffold_4690 8383893 89 - 18748788 UAGUGGCAAACUUUUUUG--CUGCGCAAGUUCGUUUUACUUUGCAA-UUUUUGUAGUU--GCUUUUGU-UUCGGUUAUUCAAAAGUUUGCCAUAA--------------- ..(((((((((((((...--..((....))(((....((...((((-((.....))))--))....))-..)))......)))))))))))))..--------------- ( -25.20, z-score = -3.33, R) >droYak2.chrX 16680917 90 - 21770863 UAGUGGCAAACUUUUGUU-GCUGCGCAAGUUCGUUUUACUUUGCAA-UUUUUGUAGUU--GCUUUUGU-UUCGGUUAUUCAAAAGUUUGCCAUAA--------------- ..((((((((((((((..-((((.((((((.......))...((((-((.....))))--))..))))-..))))....))))))))))))))..--------------- ( -29.10, z-score = -4.52, R) >droSec1.super_8 367146 91 - 3762037 UAGUGGCAAACUUUUUUGCGCUGCCCAAGUUCGUUUUACUUUGCAA-UUUUUGUAGUU--GCUUUUGU-UUCGGUUAUUCAAAAGUUUGCCAUAA--------------- ..(((((((((((((.......(((.((((.......)))).((((-((.....))))--))......-...))).....)))))))))))))..--------------- ( -24.30, z-score = -3.01, R) >droSim1.chrX_random 4644939 91 - 5698898 UAGUGGCAAACUUUUUUGCGCUGCCCAAGUUCGUUUUACUUUGCAA-UUUUUGUAGUU--GCUUUUGU-UUCGGUUAUUCAAAAGUUUGCCAUAA--------------- ..(((((((((((((.......(((.((((.......)))).((((-((.....))))--))......-...))).....)))))))))))))..--------------- ( -24.30, z-score = -3.01, R) >consensus UAGUGGCAAACUUUUUUGCGCUGCGCAAGUUCGUUUUACUUUGCAA_UUUUUGUAGUU__GCUUUUGU_UUCGGUUAUUCAAAAGUUUGCCAUAA_______________ ..(((((((((((((.......((....))..........(((((......)))))........................)))))))))))))................. (-13.41 = -13.36 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:22 2011