| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,031,142 – 18,031,261 |
| Length | 119 |
| Max. P | 0.862894 |

| Location | 18,031,142 – 18,031,239 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.36305 |
| G+C content | 0.54534 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18031142 97 + 22422827 CCUGCACGGUCGCAGCGCAGUCAUACUGG-UUCGGUAUUUUUAAGAAGAACGGUCACACUGGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAAC ............((((.(((.....)))(-((((((....(((((....((((((.....)).))))...)))))....))))))).))))....... ( -28.60, z-score = -0.25, R) >droSim1.chrX_random 148578 88 + 5698898 --------CCUGCUG-GCAGUCAUACUAG-GACGGUAUUUUUCAGCAGUACGGCCACACUAGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAAC --------(((.(((-(........))))-.).))......((((((((.(((((.....(((......)))......))))).))))))))...... ( -30.90, z-score = -2.24, R) >droSec1.super_8 340857 88 + 3762037 --------CCUGCUG-GCAGUCAUACUAG-GACGGUAUUUUUCAGCAGUACGGCCACACUAGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAAC --------(((.(((-(........))))-.).))......((((((((.(((((.....(((......)))......))))).))))))))...... ( -30.90, z-score = -2.24, R) >droYak2.chrX 16654160 98 + 21770863 CCUGCUGAGUCGUGUCGCGGUCAUACUGGCGGCGGUAUUUUUCAGCAGGACGGCCACACUGAUAUGCACGCUUAACAGGGGCAAACUGCUGAUUUAAC (((((((((..((..(((.(((.....))).)))..))..)))))))))...(((...(((.((.(....).)).))).)))................ ( -36.50, z-score = -2.60, R) >droEre2.scaffold_4690 8359199 98 + 18748788 CCUUUUGGGCCGUGGCGCGGUCAUACUGGCGGCGGUAUUAUUCAGCAGGACGGCCACACUGGCCUGCACGCUUAACAGGGGCUAACUGCUGAUUUAAC ((....))(((((.((.(((.....))))).))))).....(((((((...((((.....)))).....((((.....))))...)))))))...... ( -37.90, z-score = -1.96, R) >consensus CCU_____GCCGCGGCGCAGUCAUACUGG_GACGGUAUUUUUCAGCAGGACGGCCACACUGGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAAC .........((((...))))..((((((....))))))...(((((((..(((((.....(((......)))......)))))..)))))))...... (-19.42 = -20.06 + 0.64)
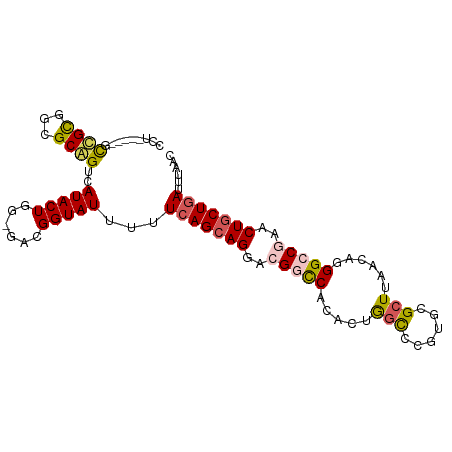
| Location | 18,031,171 – 18,031,261 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 84.84 |
| Shannon entropy | 0.25409 |
| G+C content | 0.46182 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -17.65 |
| Energy contribution | -18.49 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
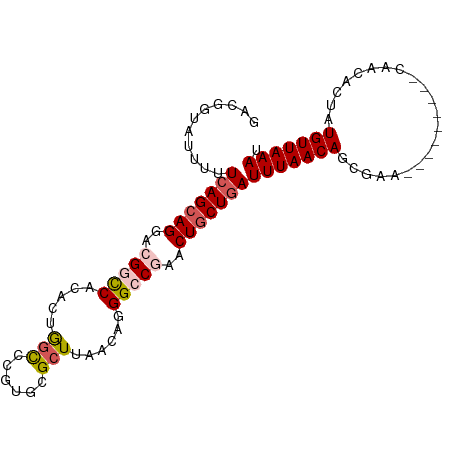
>dm3.chrX 18031171 90 + 22422827 UUCGGUAUUUUUAAGAAGAACGGUCACACUGGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGAA---------CAACACUAUGUUAAAU ((((((....(((((....((((((.....)).))))...)))))....)))))).(((((......)))))((---------((......)))).... ( -23.00, z-score = -1.51, R) >droSim1.chrX_random 148598 90 + 5698898 GACGGUAUUUUUCAGCAGUACGGCCACACUAGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGAA---------CAACACUAUGUUAAAU ...........((((((((.(((((.....(((......)))......))))).))))))))(((((((.....---------........))))))). ( -30.02, z-score = -3.63, R) >droSec1.super_8 340877 90 + 3762037 GACGGUAUUUUUCAGCAGUACGGCCACACUAGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGAA---------CAACACUAUGUUAAAU ...........((((((((.(((((.....(((......)))......))))).))))))))(((((((.....---------........))))))). ( -30.02, z-score = -3.63, R) >droYak2.chrX 16654190 99 + 21770863 GGCGGUAUUUUUCAGCAGGACGGCCACACUGAUAUGCACGCUUAACAGGGGCAAACUGCUGAUUUAACAGCGAAAAGCCAGAAUAACACUAUGUUAAAU (((.((.....(((((((....(((...(((.((.(....).)).))).)))...)))))))....))........)))....((((.....))))... ( -25.00, z-score = -1.78, R) >droEre2.scaffold_4690 8359229 99 + 18748788 GGCGGUAUUAUUCAGCAGGACGGCCACACUGGCCUGCACGCUUAACAGGGGCUAACUGCUGAUUUAACAGCGAAAAGCCAAAAUAACACUAUGUUAAAU (((.((.(((.(((((((...((((.....)))).....((((.....))))...)))))))..)))..)).....)))....((((.....))))... ( -26.70, z-score = -1.39, R) >consensus GACGGUAUUUUUCAGCAGGACGGCCACACUGGCCCGUGCGCUUAACAGGGCCGAACUGCUGAUUUAACAGCGAA_________CAACACUAUGUUAAAU ...........(((((((..(((((.....(((......)))......)))))..)))))))(((((((......................))))))). (-17.65 = -18.49 + 0.84)
| Location | 18,031,171 – 18,031,261 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 84.84 |
| Shannon entropy | 0.25409 |
| G+C content | 0.46182 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
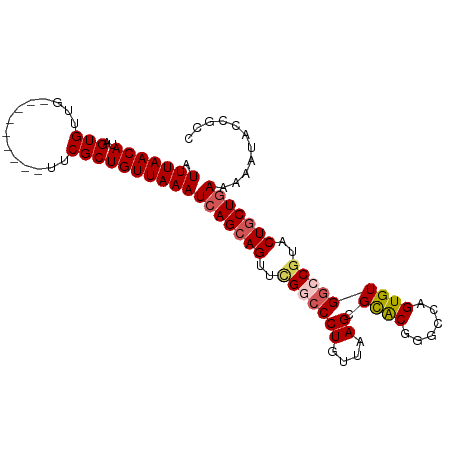
>dm3.chrX 18031171 90 - 22422827 AUUUAACAUAGUGUUG---------UUCGCUGUUAAAUCAGCAGUUCGGCCCUGUUAAGCGCACGGGCCAGUGUGACCGUUCUUCUUAAAAAUACCGAA ......((((.(....---------...((((......)))).....((((((((.....))).)))))).))))........................ ( -20.80, z-score = -0.53, R) >droSim1.chrX_random 148598 90 - 5698898 AUUUAACAUAGUGUUG---------UUCGCUGUUAAAUCAGCAGUUCGGCCCUGUUAAGCGCACGGGCUAGUGUGGCCGUACUGCUGAAAAAUACCGUC .(((((((..(((...---------..))))))))))((((((((.(((((((....)).((((......))))))))).))))))))........... ( -32.40, z-score = -3.19, R) >droSec1.super_8 340877 90 - 3762037 AUUUAACAUAGUGUUG---------UUCGCUGUUAAAUCAGCAGUUCGGCCCUGUUAAGCGCACGGGCUAGUGUGGCCGUACUGCUGAAAAAUACCGUC .(((((((..(((...---------..))))))))))((((((((.(((((((....)).((((......))))))))).))))))))........... ( -32.40, z-score = -3.19, R) >droYak2.chrX 16654190 99 - 21770863 AUUUAACAUAGUGUUAUUCUGGCUUUUCGCUGUUAAAUCAGCAGUUUGCCCCUGUUAAGCGUGCAUAUCAGUGUGGCCGUCCUGCUGAAAAAUACCGCC .(((((((..(((..(........)..))))))))))(((((((..((((((((.((.(....).)).))).).)).))..)))))))........... ( -23.00, z-score = -0.93, R) >droEre2.scaffold_4690 8359229 99 - 18748788 AUUUAACAUAGUGUUAUUUUGGCUUUUCGCUGUUAAAUCAGCAGUUAGCCCCUGUUAAGCGUGCAGGCCAGUGUGGCCGUCCUGCUGAAUAAUACCGCC ..........(((((((((.((((..(.((((......)))).)..)))).......((((....((((.....))))....))))))))))))).... ( -29.40, z-score = -2.00, R) >consensus AUUUAACAUAGUGUUG_________UUCGCUGUUAAAUCAGCAGUUCGGCCCUGUUAAGCGCACGGGCCAGUGUGGCCGUACUGCUGAAAAAUACCGCC .(((((((..(((..............))))))))))(((((((..(((((((....)).((((......)))))))))..)))))))........... (-18.76 = -19.36 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:15 2011