| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,018,383 – 18,018,475 |
| Length | 92 |
| Max. P | 0.998094 |

| Location | 18,018,383 – 18,018,475 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.08 |
| Shannon entropy | 0.47139 |
| G+C content | 0.44742 |
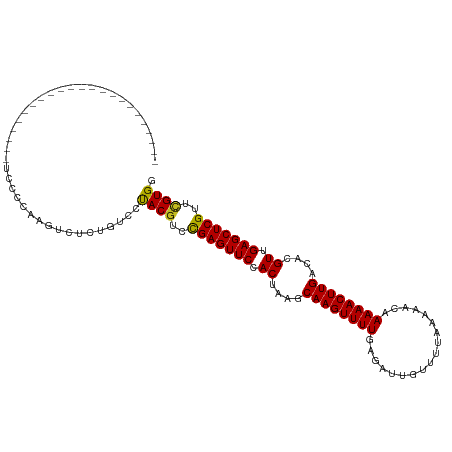
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.52 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
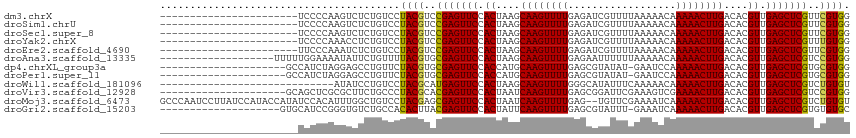
>dm3.chrX 18018383 92 + 22422827 -----------------------UCCCCAAGUCUCUGUCCUACGUCCGAGUUCCACUAAGCAAGUUUUGAGAUCGUUUUAAAAACAAAAACUUGACACGUUGAGCUCGUUCGUGG -----------------------................(((((..(((((((.((....((((((((..................))))))))....)).)))))))..))))) ( -25.27, z-score = -3.14, R) >droSim1.chrU 15519356 92 + 15797150 -----------------------UCCCCAAGUCUCUGUCCUACGUCCGAGUUCCACUAAGCAAGUUUUGAGAUCGUUUUAAAAACAAAAACUUGACACGUUGAGCUCGUUCGUGG -----------------------................(((((..(((((((.((....((((((((..................))))))))....)).)))))))..))))) ( -25.27, z-score = -3.14, R) >droSec1.super_8 326582 92 + 3762037 -----------------------UCCCCAAGUCUCUGUCCUACGUCCGAGUUCCACUAAGCAAGUUUUGAGAUCGUUUUAAAAACAAAAACUUGACACGUUGAGCUCGUUCGUGG -----------------------................(((((..(((((((.((....((((((((..................))))))))....)).)))))))..))))) ( -25.27, z-score = -3.14, R) >droYak2.chrX 16642317 92 + 21770863 -----------------------UCCCCAAACCUCUGUCCUACGUCCGAGUUCCACUAAGCAAGUUUUGAGAUCGUUUUAAAAACAAAAACUUGACACGUUGAGCUCGUUUGUGG -----------------------................(((((..(((((((.((....((((((((..................))))))))....)).)))))))..))))) ( -23.77, z-score = -2.97, R) >droEre2.scaffold_4690 8346017 92 + 18748788 -----------------------UUCCCAAAUCUCUGUCCUACGUCCGAGUUCCACUAAGCAAGUUUUGAGAUCGUUUUAAAAACAAAAACUUGACACGUUGAGCUCGUUCGUGG -----------------------................(((((..(((((((.((....((((((((..................))))))))....)).)))))))..))))) ( -25.27, z-score = -3.39, R) >droAna3.scaffold_13335 1727768 96 - 3335858 -------------------UUUUUGGAAAAUAUUCUGUUUUACGUGCGAGUUCCACUAAGCAAGUUUUGAGAAUUUUUUAAAAACAAAAACUUGACACGUUGAGCUCGUCCGUGG -------------------.....((((....))))....((((..(((((((.((....((((((((..................))))))))....)).)))))))..)))). ( -25.07, z-score = -2.36, R) >dp4.chrXL_group3a 713225 93 - 2690836 ---------------------GCCAUCUAGGAGCCUGUUCUACGUGCGAGUUCCACCAUGCAAGUUUUGAGCGUAUAU-GAAUCCAAAAACUUGACACGUUGAGCUCGUGCGUGG ---------------------......(((....)))..(((((..(((((((.((....((((((((..........-.......))))))))....)).)))))))..))))) ( -29.93, z-score = -2.37, R) >droPer1.super_11 2701278 93 + 2846995 ---------------------GCCAUCUAGGAGCCUGUUCUACGUGCGAGUUCCACCAUGCAAGUUUUGAGCGUAUAU-GAAUCCAAAAACUUGACACGUUGAGCUCGUGCGUGG ---------------------......(((....)))..(((((..(((((((.((....((((((((..........-.......))))))))....)).)))))))..))))) ( -29.93, z-score = -2.37, R) >droWil1.scaffold_181096 12029392 86 + 12416693 -----------------------------AUAUCCUGUCCUACGCAUGAGUUCCACUAAGCAAGUUUUGGGCAUAUUUCAAAAACAAAAACUUGACACGUUGAGCUCGUCUGUGU -----------------------------...........((((.((((((((.((....((((((((.(..............).))))))))....)).)))))))).)))). ( -20.24, z-score = -1.65, R) >droVir3.scaffold_12928 6453147 94 - 7717345 ---------------------GCAGCUCGCGCUUCUGCCCUACGCACGAGUUCCACUAAUCAAGUUUUGAGCGGAUUCGAAAGUCGAAAACUUGACACGUUGAGCUCGUCCGUGG ---------------------((((.........)))).(((((.((((((((.((...(((((((((..((..........))..)))))))))...)).)))))))).))))) ( -31.30, z-score = -1.82, R) >droMoj3.scaffold_6473 13724283 113 + 16943266 GCCCAAUCCUUAUCCAUACCAUAUCCACAUUUGGCUGUCCUACGAGCGAGUUCCACUAAUCAAGUUUUGAG--UGUUCGAAAAUCAAAAACUUGACACGUUGAGCUCGUCUGUGU (((.(((.....................))).))).....((((..(((((((.((...(((((((((..(--(........))..)))))))))...)).)))))))..)))). ( -26.70, z-score = -2.41, R) >droGri2.scaffold_15203 8445751 94 + 11997470 --------------------GUGCAUCCGGGUGUCUGCCACACUUACGAGUUCCACUAUUCAAGUUUUGAGCGUAUUU-GAAAUCAAAAACUUGACACGUUGAGCUCGUGUGUGC --------------------(..(((..((((((.....))))))((((((((.((...(((((((((((.((....)-)...)).)))))))))...)).)))))))))))..) ( -29.90, z-score = -2.38, R) >consensus _______________________UCCCCAAGUCUCUGUCCUACGUCCGAGUUCCACUAAGCAAGUUUUGAGAUUGUUUUAAAAACAAAAACUUGACACGUUGAGCUCGUUCGUGG ........................................((((..(((((((.((....((((((((..................))))))))....)).)))))))..)))). (-22.75 = -22.52 + -0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:11 2011